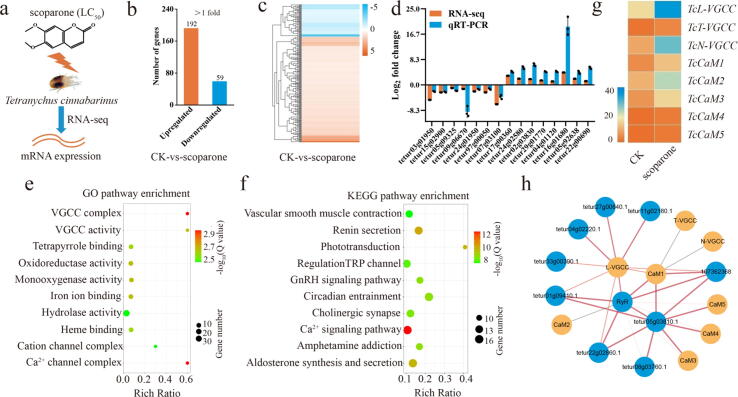
Fig. 2.
RNA-seq analysis revealed candidate targets for scoparone against T. cinnabarinus. a Diagram of the candidate targets for scoparone against mites as determined by RNA-seq. b Distributions of the differentially expressed genes in T. cinnabarinus in response to scoparone. c Heat map of significantly differentially expressed genes in the scoparone group vs the control group. d qRT-PCR validation of 15 differentially expressed genes identified by RNA-seq. e, f Top 10 enriched GO (e) and KEGG (f) pathways of the differentially expressed genes. The “rich ratio” is defined as the ratio of the number of differentially expressed genes enriched in the pathway to the total number of genes enriched in the same pathway. g Heat map of the candidate target genes involved in the Ca2+ signalling pathway. h PPI network generated from a STRING analysis. The size and colour depth of each straight line are positively correlated with the degree of connectedness with other proteins.