Figure 3.
Quantification of CSNK2B transcript and encoded protein product along with cellular localization in LCLs
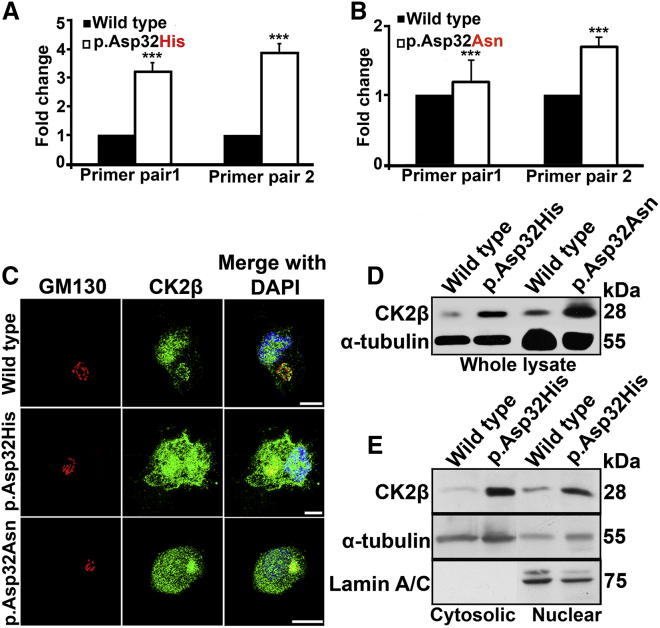
(A) Quantitative real-time PCR data showing 3-fold increased expression of CSNK2B mRNA in patient (GenBank: NM_001320.7:c.94G>C (p.Asp32His)) as compared with wild type. CSNK2B mRNA amounts were quantified relative to control sample. ∗∗∗p = 0.0001 calculated by Student’s t test. n = 3; error bars represent standard deviation (SD).
(B) Graph shows 2-fold increased expression of CSNK2B mRNA of subject (GenBank: NM_001320.7:c.94G>A (p.Asp32Asn)) resulting from amplifying a second set of primers as compared with wild type. ∗∗∗p = 0.0001 calculated by Student’s t test. n = 3; error bars represent SD.
(C) Confocal microscopy images showing increased amount of CK2β (green) in nuclei and in cytoplasm of subject-derived (NP_001311.3; p.Asp32His and NP_001311.3; p.Asp32Asn) LCLs as compared with wild type. GM130 (red) serves as marker of Golgi apparatus, and DAPI (blue) indicates staining of nucleus. Scale bar: 10 μm.
(D) Immunoblotting shows an increased amount of CK2β in whole-cell lysates obtained from mutant (NP_001311.3; p.Asp32His and NP_001311.3; p.Asp32Asn) LCLs versus wild type. α-Tubulin is used as loading controls.
(E) Immunoblots show an increased amount of CK2β in cytosolic and nuclear fraction in NP_001311.3; p.Asp32His mutant versus wild type. Loading controls are α-tubulin and lamin A/C, for cytosol and nucleus, respectively.