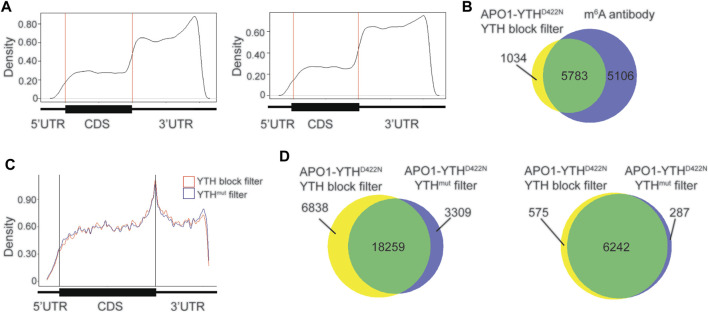
FIGURE 4.
Blocking with the YTH domain minimizes false positives with in vitro DART-seq. (A) Metagene analysis of C-to-U editing sites in mRNAs identified with in vitro DART-seq using the APO1-YTHD422N protein after pre-incubation with the YTH domain (left) or the APO1-YTHmut protein (right). (B) Venn diagram showing the overlap between methylated RNAs identified by in vitro DART-seq with APO1-YTHD422N filtered by YTH blocking and methylated RNAs identified by antibody-based methods (Meyer et al., 2012; Schwartz et al., 2014; Lichinchi et al., 2016). (C) Metagene analysis showing the distribution of C-to-U editing sites in mRNAs after filtering of in vitro DART-seq data against by YTH blocking (blue) or by APO1-YTHmut (red). (D) Venn diagram of C-to-U edit sites induced by in vitro DART-seq with APO1-YTHD422N, filtered by use of either YTH blocking or APO1-YTHmut (left). Venn diagram of methylated RNA identified by in vitro DART-seq with APO1-YTHD422N, filtered by use of either YTH blocking or APO1-YTHmut (right).