Summary
Loss of differentiated cells to tissue damage is a hallmark of many diseases. In slow turnover tissues, long-lived differentiated cells can re-enter the cell cycle or transdifferentiate to another cell type to promote repair. Here, we show that in a high turnover tissue, severe damage to the differentiated compartment induces progenitors to transiently acquire a unique transcriptional and morphological post-mitotic state. We highlight this in an acute villus injury model in the intestine, where we identified a population of progenitor-derived cells that covered injured villi. These atrophy-induced villus epithelial cells (aVECs) were enriched for fetal markers, yet were differentiated and lineage-committed. We further established a role for aVECs in maintaining barrier integrity through the activation of YAP. Notably, loss of YAP activity led to impaired villus regeneration. Thus, we define a key repair mechanism involving the activation of a fetal-like program during injury-induced differentiation, a process we term “adaptive differentiation”.
Introduction
As differentiated cells provide structure and function to tissues, these cells must be quickly replenished in response to injury. In the liver and pancreas, the initial phase of tissue regeneration relies on the plasticity of surviving mature cells (Kopp et al., 2016). These cells can undergo dedifferentiation or transdifferentiation, a concept referred to as “adaptive reprogramming” (Jessen et al., 2015). In the skin and intestine, the survival and regenerative capacity of existing pools of stem and progenitor cells are critical components of repair (Blanpain and Fuchs, 2014). Missing in these systems is an understanding of how stem and progenitor cells provide immediate structural and functional support to damaged tissues.
The mouse small intestine is an excellent system to explore the dynamic regulation of cell turnover during injury-induced repair. Distinct zones of proliferation and differentiation are organized into crypt-villus units. Crypts contain highly active intestinal stem cells (ISCs) and transit-amplifying (TA) cells; villi are lined with differentiated epithelial cells replaced every 3–5 days (Barker et al., 2007). Together, intestinal epithelial cells (IECs) must balance nutrient absorption with their role as a barrier to the harsh external environment.
Breakdown of the intestinal barrier is observed in a variety of pathologies, often triggered by damage to villus epithelial cells (VECs). Enteric viruses, hypoxic injury, endoplasmic reticulum (ER) stress, and excessive inflammation can all cause widespread VEC death (Di Sabatino et al., 2003; Hinnebusch et al., 2002; Kaser et al., 2008; Moss et al., 1996; Pensaert et al., 1970; Ramig, 2004). Such rapid loss of differentiated IECs results in villus collapse and impaired nutrient absorption (Jansson-Knodell et al., 2018). Though removal of the inciting agent typically leads to tissue recovery, persistent villus atrophy can occur for unknown reasons (Lebwohl et al., 2014; Rubio-Tapia and Murray, 2010). In understanding key features of small bowel enteropathies (Crohn’s and celiac disease), multiple studies identified disease-associated IECs with marked suppression of mature epithelial characteristics, including reduced brush border enzymes, decreased microvilli length, and diminished metabolic programs (Arvanitakis, 1979; Loberman-Nachum et al., 2019; VanDussen et al., 2018). It is not clear whether these features are a consequence of epithelial dedifferentiation, incomplete maturation, or an adaptive response to injury as we currently lack systems to model these cells.
In contrast, mechanisms underlying the preservation of ISCs during small intestine damage have been extensively studied. LGR5+ ISCs fuel normal cell turnover as well as injury-induced regeneration (Metcalfe et al., 2014). When LGR5+ ISCs are lost by irradiation or genetic ablation, multiple cell types can replenish the stem cell pool (Beumer and Clevers, 2021; de Sousa and de Sauvage, 2019). While dedifferentiation of surviving progenitors is a major mechanism of repair, injury-induced ISC states play a role as well (Murata et al., 2020; Rees et al., 2020). Specifically, the Hippo-YAP pathway is a mediator of stem cell recovery through YAP’s ability to reprogram LGR5+ ISCs to a LGR5− state, expand “revival” stem cells (revSCs), and promote a fetal-like transcriptional program (Ayyaz et al., 2019; Gregorieff et al., 2015; Yui et al., 2018). However, the precise mechanism by which YAP orchestrates repair is complicated by its crosstalk with Wnt, Notch, and EGF pathways in the crypt (Hong et al., 2016). It remains unknown whether induction of the fetal program is unique to regenerative stem cells, and whether YAP is functional in other contexts of injury independent of ISC damage.
Here, we explore the aftermath of severe damage to intestinal villi. We demonstrate that collapse of the villus architecture triggers a transient epithelial cell state that occurs in Crohn’s and celiac disease. Intriguingly, these atrophy-induced villus epithelial cells (aVECs) feature a fetal-like transcriptional profile similar to regenerative stem cells but lack stem cell capacity, are differentiated, and express lineage identity markers. We also show that aVECs are primarily derived from TA cells to repair damaged villi. Importantly, we discover a central role for YAP in promoting this adaptive response. In the absence of YAP, aVEC function is impaired and barrier integrity is compromised; as a result, villus regeneration is markedly hampered. Given that our findings cannot be explained by current models of repair, as it does not involve a conversion of cell type, we coin this process “adaptive differentiation”. We propose that adaptive differentiation represents an important healing mechanism in high turnover tissues such as the intestine.
Results
Villus injury induces a disease-associated epithelial cell type
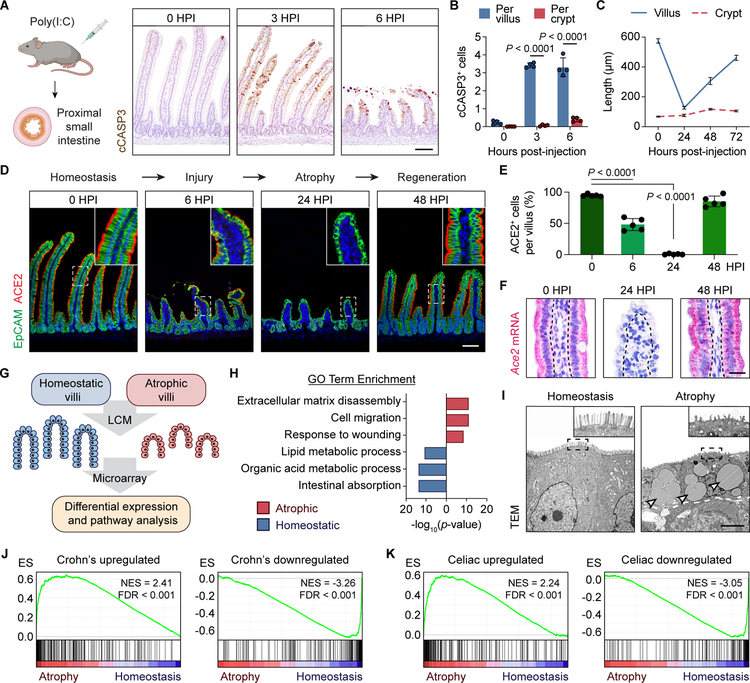
We employed a poly(I:C)-mediated damage model in mice that mimics key aspects of acute viral gastroenteritis (McAllister et al., 2013; Zhou et al., 2007). Poly(I:C) is a double-stranded RNA analog that acts directly on IECs and induces TLR3-dependent VEC death (Gunther et al., 2015; McAllister et al., 2013). In line with these studies, intraperitoneal injection of poly(I:C) rapidly triggered apoptosis of VECs in the proximal small intestine with minimal damage to crypts (Figure 1A and 1B). We observed pronounced villus atrophy by 24 hours post-injection (HPI), likely due to contraction of the myofibroblast network in the lamina propria (Figure 1C; Moore et al., 1989). This was followed by an increase in crypt depth and rapid villus regeneration. Villi reached ~50% of their original length by 48 HPI and ~75% by 72 HPI in a reproducible fashion (Figure 1C). The consistent timing and location of damage induced by poly(I:C) thus provided a unique opportunity to study the response of villi to injury.
Figure 1. Villus injury induces a disease-associated epithelial cell type.
(A and B) Immunohistochemistry (IHC) for cleaved-caspase 3 (cCASP3) (brown) at the indicated hours post-injection (HPI) of poly(I:C) (A). Average number of cCASP3+ cells across 50 villi/crypts was plotted (B). n=4 mice/group.
(C) Average villus/crypt length across 50 villi/crypts at the indicated time points based on H&E images was plotted. n=4 mice/group.
(D and E) Immunofluorescence (IF) for EpCAM (green) and ACE2 (red) at the indicated time points (D). Average number of ACE2+ cells across 30 villi was plotted (E). n=5 mice/group.
(F) RNAscope in situ hybridization for Ace2 in a representative villus at the indicated time points. Each red dot represents a single mRNA molecule.
(G) Schematic of laser-capture microdissection (LCM)-microarray experiment. Villus epithelial cells (VECs) from 4 mice/group were collected.
(H) Gene ontology (GO) analysis of the top 500 upregulated and downregulates genes in atrophy-induced VECs (aVECs) compared with homeostatic VECs.
(I) Ultrastructure of homeostatic VECs and aVECs. Dashed white line represents the epithelial-stromal border. aVECs possess shorter microvilli (inset) and contain many lipid droplets (arrowheads).
(J and K) Gene set enrichment analysis (GSEA) of genes upregulated or downregulated in Crohn’s disease (J) and celiac disease (K) in aVECs compared with homeostatic VECs. Normalized enrichment scores (NES) and false discovery rates (FDR) are shown.
All values in (B), (C), and (E) are displayed as mean ± SD. Two-way ANOVA and Sidak’s multiple comparisons test in (B). One-way ANOVA and Tukey’s multiple comparison test in (E). Bars: (A and D) 100μm; (F) 25μm; (I) 5μm. IHC, IF, RNAscope, and electron microscopy images are representative of at least 3 animals. See also Figure S1.
To assess epithelial differentiation dynamics in this model throughout the injury-repair process, we first examined the villus expression pattern of ACE2, a brush border protein and a marker of mature enterocytes, the dominant cell type on villi (Zang et al., 2020). During the injury phase (6 HPI), ACE2+ enterocytes were shed into the lumen and lost. During villus atrophy (24 HPI), a thin epithelial layer that lacked ACE2 protein and mRNA expression covered rudimentary villi in the proximal small intestine (Figure 1D–1F, S1A, and S1B). Expression of a second enterocyte marker FABP1 was similarly downregulated during the atrophy phase (Figure S1C), indicating a suppression of the normal differentiation program. These epithelial changes were transient as ACE2+ and FABP1+ columnar-shaped enterocytes quickly reappeared during the regenerative phase (>48 HPI; Figure 1D–1F and S1C).
A reduction of mature enterocyte features occurs in a variety of infectious and inflammatory small bowel pathologies (Holmes and Lobley, 1989). To determine the scope of the overall transcriptional program impacted by villus injury, we performed microarray analysis of laser-capture microdissected (LCM) epithelial cells from homeostatic and atrophic villi (Figure 1G and S1D). Atrophy-induced VECs (hereafter aVECs) possessed a distinct transcriptional state compared to homeostatic VECs (Figure S1E). Pathways related to cell migration and wound healing were induced in aVECs, whereas pathways linked to intestinal absorption and enterocyte metabolism were suppressed (Figure 1H). Consistent with these transcriptional changes, homeostatic VECs were comparatively taller with well-formed microvilli, while aVECs were smaller with poorly developed microvilli and contained extensive lipid droplets (Figure 1I).
To test the hypothesis that an aVEC-like state is present in human diseases with injured villi, we compared the transcriptional signature of aVECs in mice to bulk RNA sequencing datasets of Crohn’s and celiac disease biopsies. We found two established epithelial biomarkers of Crohn’s disease, Duox2 and Lcn2 (Csillag et al., 2007), were among the most highly upregulated genes in aVECs (Figure S1F). Gene set enrichment analysis (GSEA) showed significant correlation of a Crohn’s disease signature with the aVEC dataset (Figure 1J; Haberman et al., 2014). We also found significant correlation of two celiac disease signatures with the aVEC dataset that was dependent on active disease status (Figure 1K and S1G; Dotsenko et al., 2021; Loberman-Nachum et al., 2019). Therefore, the aVEC signature can be identified in the transcriptomes of mucosal specimens from multiple human enteropathies.
aVECs acquire a fetal-like transcriptional program
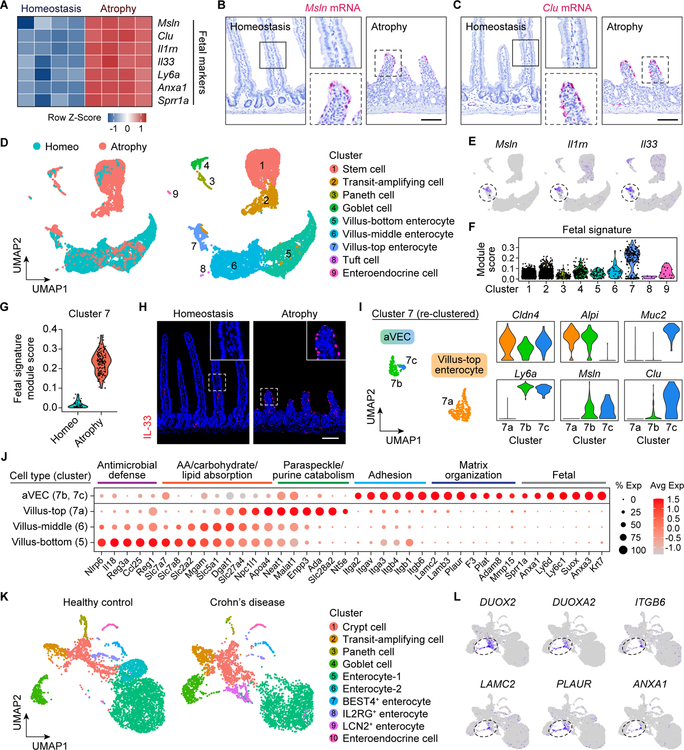
During epithelial regeneration, ISCs have been shown to express a fetal-like signature (Gregorieff et al., 2015; Nusse et al., 2018; Wang et al., 2019; Yui et al., 2018). Surprisingly, we found components of a fetal program in the aVEC transcriptome, including the expression of Msln and Clu (Figure 2A and S2A). In situ hybridization confirmed the induction of Msln and Clu mRNAs in aVECs (Figure 2B and 2C). Furthermore, GSEA demonstrated significant correlation of a fetal program with the bulk transcriptome of aVECs (Figure S2B; Mustata et al., 2013).
Figure 2. aVECs acquire a fetal-like transcriptional program.
(A) Heatmap of select fetal markers from the aVEC LCM-microarray dataset. Values were row normalized.
(B and C) RNAscope for Msln (B) and Clu (C) in the homeostatic and atrophic intestine. Bars: 100μm.
(D) UMAP visualization of intestinal epithelial cells (IECs) from the homeostatic (4,892 cells) and atrophic (3,650 cells) intestine colored by sample (left) and cell type (right). Clusters were annotated based on expression of known and top marker genes.
(E) Expression of select fetal markers was overlaid on the UMAP plot. Highest expressors were concentrated in a subset of villus-top enterocytes (cluster 7, dashed circle).
(F and G) Enrichment analysis of a fetal signature separated by cluster (F) and sample in cluster 7 (G). Each black dot represents one cell.
(H) IF for IL-33 (red) in the homeostatic and atrophic intestine. Bar: 100μm.
(I) UMAP visualization of villus-top enterocyte (cluster 7) sub-clusters colored by cell type (left). Expression of villus tip (Cldn4), cell lineage (Alpi, Muc2), and fetal (Ly6a, Msln, Clu) markers in each sub-cluster (right).
(J) Dot plot of enterocyte zonation markers from the villus-bottom to villus-top. Cell-adhesion, extracellular matrix, and fetal genes expressed by aVECs (atrophy-top cells) are shown.
(K) UMAP visualization of IECs from healthy control (4,630 cells) and pediatric Crohn’s disease (3,545 cells) patients obtained from the Gut Cell Survey. Clusters were annotated based on expression of known and top marker genes.
(L) Expression of select aVEC markers was overlaid on the UMAP plot. Highest expressors were concentrated in DUOX2+ enterocytes and goblet cells.
RNAscope and IF images are representative of at least 3 animals. See also Figure S2.
We further tested the hypothesis that aVECs possess a fetal-like state by single-cell RNA sequencing (scRNA-seq) of IECs isolated during homeostasis and poly(I:C)-induced villus atrophy (Figure S2C and S2D). Uniform manifold approximation and projection (UMAP) of the integrated dataset identified all major intestinal cell lineages, including three enterocyte clusters reflective of their position along the villus axis (Figure 2D and S2E; Moor et al., 2018). Enterocytes were particularly enriched in the homeostasis sample, while crypt-based and secretory cells were enriched in the atrophy sample (Figure S2F). Interestingly, we detected highest expression of fetal markers and enrichment of a fetal signature in the villus-top enterocyte cluster (cluster 7) (Figure 2E and 2F). In line with this, a villus tip marker CLDN4 was expressed by both aVECs and VECs near the top of homeostatic villi (Figure S2G; Tamagawa et al., 2003). However, cells possessing the fetal signature were mainly present in the atrophy sample, as demonstrated by the expression of IL-33 in aVECs but not in homeostatic VECs (Figure 2G and 2H). Given these findings, we further re-clustered cluster 7 and identified three distinct sub-clusters, two of which belonged to aVECs (cluster 7b and 7c) and the other to villus-top enterocytes (cluster 7a). Marker analysis identified aVECs as altered versions of Alpi+ enterocytes and Muc2+ goblet cells enriched for fetal markers (Figure 2I). aVECs did not express enterocyte zonation markers present in homeostatic VECs, and were instead enriched for cell adhesion, extracellular matrix, and fetal genes (Figure 2J). Following the atrophy phase, expression of a villus-top enterocyte marker NT5E was detected on regenerating villi, likely owing to restoration of the stromal niche provided by PDGFRα+ villus tip telocytes (Figure S2H; Bahar Halpern et al., 2020). Together, these results show aVECs feature a unique injury-induced enterocyte and goblet cell state absent in the normal adult intestine.
As aVECs can be defined in mice at single-cell resolution, we next examined a recent scRNA-seq atlas for Crohn’s disease (Elmentaite et al., 2020). Analysis of the epithelial cell composition in newly diagnosed Crohn’s disease patients and healthy controls yielded 10 clusters with distinct subsets of enterocytes (Figure 2K and S2I). Overall, there was a reduction in the relative abundance of absorptive cells and an increase in crypt-based and secretory cells in Crohn’s disease samples (Figure S2J). Notably, we observed an expansion of a unique LCN2+ enterocyte population in Crohn’s disease (Figure S2K). Together with a subset of Crohn’s disease-associated goblet cells, LCN2+ enterocytes expressed DUOX2 and several other aVEC markers, including ITGB6, LAMC2, PLAUR, and ANXA1 (Figure 2L and S2L). In addition, these cells were enriched for both aVEC and mouse fetal signatures (Figure S2M and S2N), suggesting an aVEC-like cell type may also be present in human enteropathies.
aVECs are short-lived differentiated cells
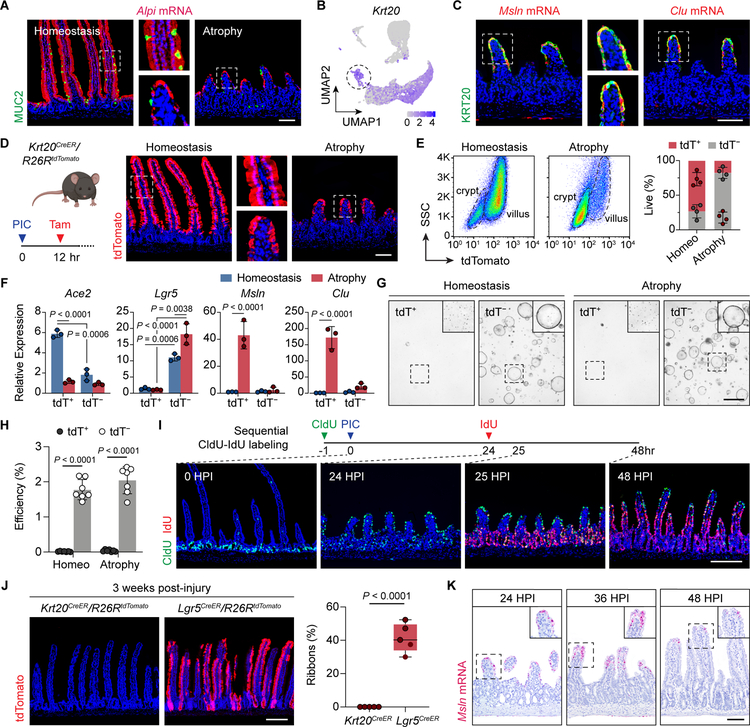
Due to the prominent fetal signature in aVECs, we next tested their stemness and lineage properties. Though aVECs lack the expression of specialized proteins associated with mature enterocyte function (ACE2 and FABP1), these cells nevertheless maintain either an enterocyte or goblet cell identity consistent with our single-cell data: Alpi mRNA and MUC2 protein were expressed in discrete cells on atrophic villi (Figure 3A). Additionally, all Msln+ and Clu+ aVECs expressed the differentiation marker KRT20 (Figure 3B and 3C). Other intestinal cell lineage markers for Paneth, enteroendocrine, and tuft cells were detected during villus atrophy at frequencies and numbers comparable to homeostatic levels (Figure S3A). Interestingly, the fetal intestine is also characterized by the absence of enterocyte protein markers with expression of KRT20, MUC2, and Alpi mRNA (Guiu et al., 2019). Overall, no difference in Wnt signaling activity was appreciated between aVECs and homeostatic VECs (Figure S3B).
Figure 3. aVECs are short-lived differentiated cells.
(A) IF/RNAscope for MUC2 (green) and Alpi (red) in the homeostatic and atrophic intestine.
(B) Expression of Krt20, an intestinal differentiation marker, was overlaid on the UMAP plot. Villus-top enterocytes (dashed circle) express Krt20.
(C) IF/RNAscope for KRT20 (green) and Msln or Clu (red) in the atrophic intestine.
(D) Validation of tamoxifen-mediated fluorescent labeling of KRT20+ cells in Krt20CreER/R26RtdTomato mice during homeostasis and atrophy. Labeled cells express tdTomato (tdT).
(E) Flow cytometry analysis of KRT20-tdT+ and KRT20-tdT− cells during homeostasis and atrophy. Representative plots (left) and relative percentage of each population (right). n=4 mice/group.
(F) qPCR analysis of indicated transcripts in sorted KRT20-tdT+ and KRT20-tdT− cells during homeostasis and atrophy. Values were normalized to the lowest expressing sample. n=3 samples/group.
(G and H) Sorted KRT20-tdT+ and KRT20-tdT− cells were cultured in Matrigel with 50% L-WRN conditioned media. Bright-field images of spheroids were taken on day 6 after plating (G). Insets were enhanced for contrast. Spheroid formation efficiency = number of spheroids on day 6 over number of seeded cells on day 0 (H). n=7 mice/group.
(I) Assessment of epithelial turnover dynamics. CldU was injected 1 h prior to poly(I:C) injection, and IdU was injected at 24 HPI. IF for CldU (green) and IdU (red) at the indicated time points.
(J) Transiently labeled tdT+ cells from Krt20CreER/R26RtdTomato and Lgr5CreER/R26RtdTomato mice were examined 3 weeks post-injury (left). Percent of ribbons across 50 crypt-villus units was quantified and plotted as a box-whisker plot (right). n=5 mice/group.
(K) RNAscope for Msln at the indicated time points.
All values in (E), (F), and (H) are displayed as mean ± SD. Two-way ANOVA and Sidak’s multiple comparisons test in (F) and (H). Unpaired t-test in (J). Bars: (A, C, D, and K) 100μm; (G) 1000μm; (I and J) 200μm. IF and RNAscope images are representative of at least 3 animals. See also Figure S3.
Clu is expressed in aVECs as we described, but is also a defining marker of revSCs, which drive epithelial regeneration after loss of LGR5+ ISCs to intestinal damage (Ayyaz et al., 2019). Given the close link between the fetal program and regenerative stem cells, we tested whether aVECs were endowed with stem cell capacity or represented a terminally differentiated cell population. We transiently labeled KRT20-expressing VECs at homeostasis and during villus atrophy by tamoxifen induction of tdTomato in Krt20CreER/R26RtdTomato mice (Figure 3D). Flow cytometry analysis of IECs demonstrated that 73% were tdTomato (tdT)+ at homeostasis compared to 18% during atrophy (Figure 3E). We validated that KRT20-tdT+ aVECs were enriched for the fetal markers Msln and Clu with reduced expression of Ace2 compared to KRT20-tdT+ homeostatic VECs (Figure 3F). To functionally test for stemness, we cultured sorted tdT+ and tdT− cells in Matrigel/L-WRN conditioned media (Miyoshi and Stappenbeck, 2013). KRT20-tdT+ aVECs and homeostatic VECs (as a control) failed to form spheroids, suggesting a lack of stem cell capacity. In contrast, KRT20-tdT− cells showed robust spheroid-forming efficiency regardless of injury as expected of crypt cells (Figure 3G and 3H).
To determine the source of aVECs, we first examined turnover dynamics during the injury-repair process by injecting the thymine analogs CldU and IdU prior to injury and during villus atrophy, respectively. These data showed TA cells migrate over damaged villi to become aVECs, which are then replaced by newly emerged IECs during villus regeneration (Figure 3I). Consistent with this finding, lineage tracing identified the recent progeny of LGR5+ ISCs as the major source of aVECs relative to LGR5+ and BMI1+ cells (Figure S3C–S3E). While we cannot exclude the possible contribution of other cell types to aVEC formation, including pre-injury villus cells (Figure S3F), our findings favor a model in which TA cells differentiate into aVECs after injury and acquire a fetal-like state. Since this program of repair occurs in the setting of normal lineage specification, we termed this process “adaptive differentiation”.
The short-lived nature of the aVEC population was further corroborated by the fact KRT20-labeled cells were absent three weeks post-injury (Figure 3J). In contrast, LGR5+ ISCs labeled prior to or immediately after injury robustly contributed to regeneration (Figure 3J and S3D). Tracking the fate of Msln+ cells by in situ hybridization showed that aVECs were replaced and sloughed off during the regenerative phase (Figure 3K). Taken together, we demonstrate that TA cells adaptively differentiate into aVECs, which transiently cover injured villi.
YAP is activated in aVECs during barrier re-establishment
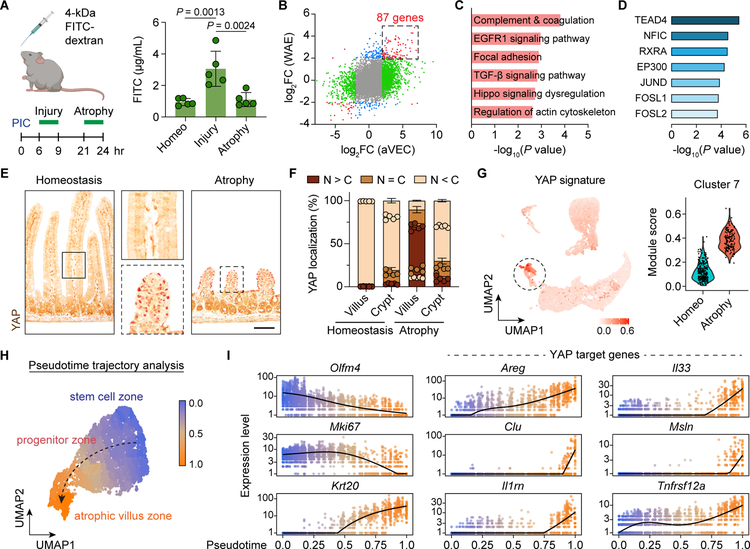
We next investigated the function of aVECs during repair and the programs that drive it. Since pathway analysis of the aVEC dataset highlighted wound healing programs, we hypothesized that these cells protect the surface of damaged villi. Serum levels of orally-gavaged FITC-dextran during the injury phase showed marked barrier leakage that later resolved during the atrophy phase (Figure 4A), revealing aVECs possess barrier restorative properties.
Figure 4. YAP is activated in aVECs during barrier re-establishment.
(A) Schematic of FITC-dextran permeability assay (left). FITC-dextran was gavaged and serum was obtained 3 h later. Serum FITC-dextran levels were measured and plotted as mean ± SD (right). n=5 mice/group. Significance was determined by one-way ANOVA and Tukey’s multiple comparison test.
(B) Fold change values for each gene from the aVEC and wound-associated epithelial (WAE) cell LCM-microarray datasets were plotted. Differentially expressed genes in only the aVEC dataset were colored green, in only the WAE dataset were colored blue, and in both datasets were colored red. 87 mRNAs were commonly enriched in both aVECs and WAE cells.
(C and D) Pathway analysis (C) and transcription factor motif analysis (D) of the aVEC-WAE shared gene set.
(E) IHC for YAP (brown) in the homeostatic and atrophic intestine. Bar: 100μm.
(F) Percent of epithelial cells with predominantly nuclear (N>C), equally nuclear and cytoplasmic (N=C), or predominantly cytoplastic (N<C) localization of YAP was quantified across 30 villi/crypts based on IF images and plotted as mean ± SD. n=5 mice/group.
(G) Expression of a YAP signature was overlaid on the UMAP plot (left) and enrichment scores were plotted for each cell in cluster 7 separated by sample (right).
(H and I) Pseudotime analysis of IECs during villus atrophy based on single cell transcriptomes. Cells were colored by progression through a pseudotime differentiation trajectory (H). Black dashed arrow indicates direction of fate progression. Single cells were plotted according to their pseudotime position and their expression level of stem cell (Olfm4), proliferation (Mki67), differentiation (Krt20), and YAP target genes (I).
To discover candidate pathways that could control the function of aVECs, we cross-compared the aVEC dataset with the transcriptome of wound-associated epithelial (WAE) cells that mediate the initial step of repair following colonic injury (Miyoshi et al., 2012; Seno et al., 2009). This analysis defined 87 genes commonly induced in both cell types, many of which were fetal markers (Figure 4B; Table S1). Shared pathways included coagulation, EGFR, Hippo, and focal adhesion (Figure 4C). Importantly, TEAD4 and AP-1, which act downstream of Hippo signaling and synergize to regulate gene expression (Zanconato et al., 2015), were among the top transcription factors whose motifs were enriched in this signature (Figure 4D). Thus, we hypothesized that barrier restoration after villus injury required the Hippo signaling pathway.
In the context of crypt injury induced by dextran sodium sulfate (DSS) or irradiation, Hippo signaling is essential for proper epithelial regeneration through activation of the transcriptional regulator YAP (Cai et al., 2010; Gregorieff et al., 2015). To assess whether YAP is involved in aVECs during villus repair, we compared the localization of YAP between the homeostatic and atrophic intestine. At baseline, YAP was largely cytoplasmic throughout the epithelium. In contrast, YAP was predominantly nuclear in aVECs, reflecting an activated state, and largely retained cytoplasmic localization in the crypt after injury (Figure 4E and 4F). In line with these observations, YAP signature genes were almost exclusively expressed by atrophy-associated cluster 7 cells (aVECs) (Figure 4G). Additionally, YAP staining colocalized with Msln and Clu mRNA expression (Figure S4A and S4B). These results suggest that YAP may act as a sensor of tissue integrity in multiple epithelial compartments in the gut, including villi.
We next mapped single cells in the villus atrophy sample along a pseudotime trajectory using Monocle 3 (Cao et al., 2019), with cells expressing high levels of ISC markers as the starting point (Figure 4H). This unsupervised approach revealed induction of YAP target genes over the course of epithelial differentiation, especially upon exit of the progenitor cell zone (Figure 4I). YAP activation has been previously reported in “resealing epithelial cells” in the ischemia-reperfusion damage model (Takeda and Kiyokawa, 2017). In the colonic biopsy injury system, WAE cells were comprised of primitive-appearing colonocytes and goblet cells with pronounced YAP nuclear staining, high Clu expression, and significant enrichment of the YAP program (Figure S4C–S4H). Progenitors were also a major source of WAE cells and resealing epithelial cells (Figure S4I; Seno et al., 2009; Takeda and Kiyokawa, 2017). These findings indicate that YAP activation may be a general feature of adaptively differentiated IECs.
We then examined the expression of stromal-derived factors previously implicated in YAP activation, revSC expansion, and WAE cell production to test for association with aVECs. These factors included PTGS2, the gp130 ligands IL-6 and IL11, and prostaglandin E2 (PGE2) (Kim et al., 2017; Manieri et al., 2012; Miyoshi et al., 2017; Roulis et al., 2020; Taniguchi et al., 2015). qPCR analysis showed a robust increase in Ptgs2, Il6, and Il11 transcript levels in the atrophic intestine (Figure S4J). PGE2 was also elevated specifically during villus atrophy (Figure S4K). Notably, while Ptgs2 and Il11 were lowly expressed in the homeostatic intestine, these transcripts were highly induced within the stroma of atrophic villi (Figure S4L). Therefore, a stromal environment conducive to YAP activation is induced during aVEC formation.
YAP is critical for barrier function and epithelial integrity
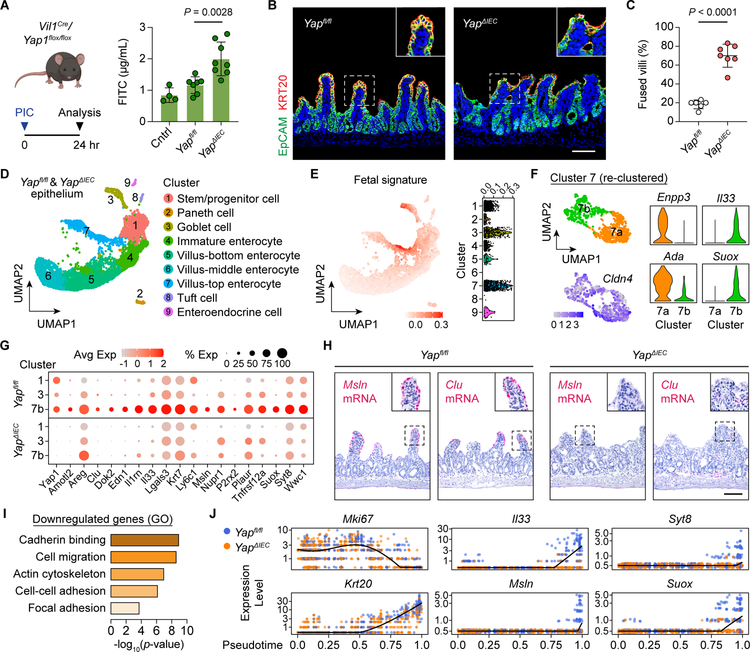
We next tested the role of YAP in barrier restoration using Vil1Cre/Yapfl/fl (YapΔIEC) mice (Figure S5A). As previously reported, deletion of Yap did not perturb intestinal homeostasis (Cai et al., 2010). While the level of IEC death induced by poly(I:C) was comparable between Yapfl/fl and YapΔIEC mice (Figure S5B and S5C), intestinal permeability to FITC-dextran was elevated in YapΔIEC mice during villus atrophy (Figure 5A), demonstrating a compromised epithelial barrier. In addition, histological analysis revealed significantly more atrophic villi that were fused together in YapΔIEC mice compared to littermate Yapfl/fl controls (Figure 5B and 5C).
Figure 5. YAP is critical for barrier function and epithelial integrity.
(A) FITC-dextran was gavaged at 24 HPI and serum was obtained 3 h later from Yapfl/fl and YapΔIEC mice. Serum FITC-dextran levels were measured and plotted as mean ± SD (right). Control (cntrl) were Yapfl/fl mice without poly(I:C) injection. n=4–8 mice/group. Significance was determined by one-way ANOVA and Tukey’s multiple comparison test.
(B and C) IF for EpCAM (green) and KRT20 (red) in the atrophic intestine from Yapfl/fl and YapΔIEC mice (B). Bar: 100μm. Percent of atrophic villi that were fused to other villi was quantified across 50 villi and plotted as mean ± SD (C). n=6–7 mice/group. Significance was determined by unpaired t-test.
(D) UMAP visualization of IECs from Yapfl/fl (3,571 cells) and YapΔIEC (4,013 cells) mice colored by cell type. Clusters were annotated based on expression of known and top marker genes.
(E) Expression of a fetal signature was overlaid on the UMAP plot (left) and enrichment scores were plotted for each cell separated by cluster (right).
(F) UMAP visualization of villus-top enterocyte (cluster 7) sub-clusters colored by cell type (left top) and relative Cldn4 expression (left bottom). Expression of villus-top (Enpp3, Ada) and fetal (Il33, Suox) markers in each sub-cluster (right).
(G) Dot plot of YAP target genes in stem/progenitor cells (cluster 1), goblet cells (cluster 3), and atrophy-induced enterocytes (cluster 7b) separated by genotype.
(H) RNAscope for Msln and Clu in the atrophic intestine from Yapfl/fl and YapΔIEC mice. Bar: 100μm.
(I) GO analysis of downregulated genes in YapΔIEC cells compared with Yapfl/fl cells.
(J) Single Yapfl/fl cells (blue) and YapΔIEC cells (orange) were plotted according to their pseudotime position and their expression level of proliferation (Mki67), differentiation (Krt20), and YAP target genes.
IF and RNAscope images are representative of at least 3 animals. See also Figure S5.
To elucidate the transcriptional program regulated by YAP specifically in aVECs during repair, we performed scRNA-seq of IECs from the proximal intestine with roughly equivalent amount of the atrophic and distal non-atrophic regions of the gut to capture the full spectrum of cell states in Yapfl/fl and YapΔIEC mice (Figure S5D–S5F). UMAP identified all major intestinal cell lineages and revealed a similar clustering pattern between the two genotypes (Figure 5D and S5G). Consistent with our prior scRNA-seq analysis, the fetal program emerged most prominently in the villus-top enterocyte (cluster 7) and goblet cell (cluster 3) clusters (Figure 5E). Likewise, the villus-top enterocyte cluster could be further re-clustered into two distinct Cldn4-expressing clusters, one corresponding to Enpp3+Ada+ homeostatic villus-top enterocytes (cluster 7a) and the other to Il33+Suox+ atrophy-induced enterocytes (cluster 7b) (Figure 5F).
Homing in on the clusters representing stem/progenitor cells (cluster 1) and fetal program-enriched IECs (clusters 3 and 7b), we performed comparative analysis between Yapfl/fl and YapΔIEC cells for each cluster. Reduction of the YAP program was greatest in atrophy-induced enterocytes (Figure 5G and S5H). In situ hybridization validated the loss of Msln and Clu expression in aVECs in YapΔIEC mice (Figure 5H). Interestingly, the expression of Areg was not affected despite being a YAP target gene in regenerating crypts (Figure S5I; Gregorieff et al., 2015). Top pathways impacted by Yap deletion included cell-cell adhesion, cell migration, actin cytoskeleton, and focal adhesion, similar to those regulated by YAP in intestinal organoids (Figure 5I and S5J). Reconstruction of single cells along a pseudotime trajectory confirmed the requirement for YAP in invoking the adaptive differentiation program (Figure 5J and S5K). Together, these data establish a critical role for YAP in maintaining epithelial barrier integrity.
Upon damage to ISCs, YAP inhibits Wnt signaling, leading to loss of canonical ISC markers, in order to prevent excessive Paneth cell differentiation (Gregorieff et al., 2015). While we observed a minor increase in the expression of Wnt, stem cell, and Paneth cell genes upon Yap deletion, this was not unique to the stem/progenitor cell cluster (Figure S5L). Quantification of ISCs and Paneth cell numbers by immunostaining revealed no difference between Yapfl/fl and YapΔIEC crypts during villus atrophy (Figure S5M). The number of Ki-67+ proliferating crypt cells also did not significantly differ (Figure S5N). These results are consistent with the lack of ISC damage in our model and the primary role of YAP in mediating adaptive differentiation.
YAP deficiency impairs villus regeneration
Given the mechanistic link between YAP activation and the fetal-like epithelial conversion seen in DSS-induced colitis (Yui et al., 2018), we examined the extent to which YAP controls the aVEC state. In determining which of the fetal (Mustata et al., 2013) and YAP signature (Gregorieff et al., 2015) genes were aVEC-specific and/or YAP-dependent in the poly(I:C) injury model, we discovered that many of the overlapping genes between the two datasets were uniquely expressed by aVECs. Importantly, only ~40% of these aVEC markers were impacted by Yap deletion (Figure S6A; Table S2). These findings reinforce the idea that YAP serves to positively modulate aVEC function rather than acting as a reprogramming factor.
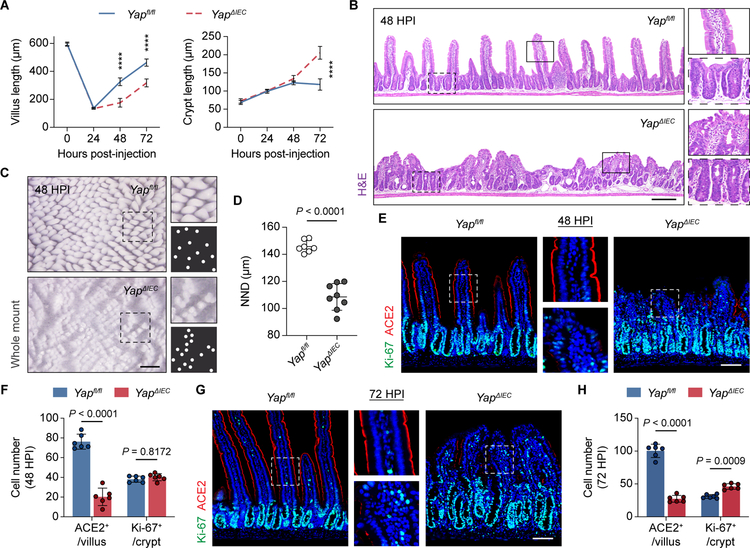
To assess the consequence of having dysfunctional aVECs in the setting of Yap deficiency, we examined the regenerative capacity of villi following injury. Notably, compared to Yapfl/fl controls, villus regeneration was hampered in YapΔIEC mice at 48 and 72 HPI. This was accompanied by compensatory expansion of the crypt at 72 HPI (Figure 6A). Histological analysis at 48 HPI during the regenerative phase revealed striking aggregates of fused and stunted villi in YapΔIEC mice (Figure 6B). Whole mount imaging and spatial distribution analysis showed that villus regeneration occurred in a more clustered pattern in the absence of Yap at 48 HPI (Figure 6C and 6D). While our collective data position YAP’s activity primarily in the villus compartment in our model, we were unable to specifically delete Yap in differentiated cells using Krt20CreER/Yapfl/fl mice to confirm this (Figure S6B). This is likely because YAP is produced in the crypt and the protein persists in the villus epithelium (Camargo et al., 2007).
Figure 6. YAP deficiency impairs villus regeneration.
(A) Average villus/crypt length across 50 villi/crypts for each time point based on H&E images was plotted. n=6–8 mice/group. ****p < 0.0001.
(B) H&E images of the regenerating intestine at 48 HPI from Yapfl/fl and YapΔIEC mice.
(C and D) Whole mount luminal view of regenerating villi at 48 HPI from Yapfl/fl and YapΔIEC mice (C). White dots represent individual villus tips. The distance between a villus and its closest neighboring villus (nearest neighbor distance, NND) was calculated based on the whole-mount images, and the average value was plotted (D). n=7–8 mice/group.
(E-H) IF for Ki-67 (green) and ACE2 (red) in the regenerating intestine at 48 HPI (E) and 72 HPI (G) from Yapfl/fl and YapΔIEC mice. Average number of Ki-67+ and ACE2+ cells across 30 villi/crypts at 48 HPI (F) and 72 HPI (H) was plotted. n=6 mice/group.
All values in (A), (D), (F), and (H) are displayed as mean ± SD. Unpaired t-test with Welch’s correction in (A). Unpaired t-test in (D). Two-way ANOVA and Sidak’s multiple comparisons test in (F) and (H). Bars: (B) 200μm; (C) 500μm; (E and G) 100μm. H&E, whole mount, and IF images are representative of at least 3 animals. See also Figure S6.
Further characterization of the stunted YapΔIEC villi showed that the epithelium was in a persistent maladapted aVEC state (Figure S6C). Differentiation was impaired with reduced ACE2+ enterocytes and abnormal-appearing goblet cells on regenerating villi (Figure 6E, 6F, and S6D). By 72 HPI, YapΔIEC villi continued to show impaired recovery with diminished ACE2 expression, and the crypts at this time point were hyperproliferative (Figure 6G and 6H). By one-week post-injury, YapΔIEC mice had largely restored their villus architecture with signs of normal epithelial differentiation (Figure S6E and S6F). In addition to the elevated crypt response, compensatory stromal responses may aid with villus regeneration as demonstrated by the heightened Ereg expression in YapΔIEC mice (Figure S6G and S6H; Gregorieff et al., 2015). Thus, YAP’s early role in villus repair can profoundly impact the regenerative outcome of villi.
Discussion
Severe tissue injury triggers unique cellular states not observed under normal physiological conditions. Here, we adopted a poly(I:C) injury mouse model to delineate the steps involved in repairing the villus structure. We found that following villus injury, TA cells rapidly covered the surface of atrophic villi and transiently differentiated into aVECs—a post-mitotic, lineage-committed cell state with a fetal-like transcriptional program. Functionally, aVECs were important for re-establishing the intestinal barrier through activation of the Hippo signaling regulator YAP, which was previously shown to promote wound healing behaviors in other cell types through modification of cell mechanics, cell adhesion, and actin cytoskeleton (Calvo et al., 2013; Nardone et al., 2017; Neto et al., 2018). Restoration of the epithelial barrier was followed by crypt expansion, villus regeneration, and return of a normal differentiation program.
Historically, rapid resealing of the mucosal barrier after disruption of the gastrointestinal tract has been referred to as “restitution” (Lacy, 1988; Sturm and Dignass, 2008). The term was first used to describe the repair of the amphibian gastric lining after severe hypertonic chemical injury (Svanes et al., 1982). Work that followed recognized that the repairing epithelium went through a phase where the cell shape changed dramatically, implying that tissue repair did not simply involve the production of new fully differentiated cells. In addition, restitution was initially thought to involve a partial dedifferentiation step (Sturm and Dignass, 2008); however, the molecular features of these injury-induced changes remained largely uncharacterized. Using a biopsy injury system, we previously described a colonic WAE cell population that repairs wounds after extensive crypt loss (Miyoshi et al., 2017; Seno et al., 2009). However, the paucity of these cells in this model, the severe disruption of mucosal architecture caused by the injury, and the lack of high-resolution transcriptomic analysis limited our ability to define the lineage properties of WAE cells and the mechanisms responsible for their barrier function.
Using a small intestine injury model, we defined an aVEC population that repairs injured villi. Morphologically, aVECs were distinct from mature IECs. They were primitive in their appearance with a short cuboidal morphology and diminished brush border, and they resembled surface epithelial cells associated with intestinal disease (Kent and Moon, 1973; Kerzner et al., 1977; Schuffler and Chaffee, 1979). By transcriptomics, we identified the presence of aVEC-like cells in Crohn’s and celiac disease samples, further highlighting the relevance of our study.
Along with recently identified regenerative stem cells (Ayyaz et al., 2019; Wang et al., 2019), aVECs are another population of cells in the gut that possess a fetal-like, YAP-activated profile. While studies so far associate this damage-induced state with stemness and regeneration (Beumer and Clevers, 2021; Nusse et al., 2018; Sprangers et al., 2021), our data suggest that it can also be observed under certain contexts of epithelial differentiation. It is therefore likely that proximity to niche factors in the crypt is crucial for determining stem cell function (Fuchs et al., 2004; Gehart and Clevers, 2019). However, no current models of repair adequately explain the mechanism for aVEC formation. While fetal-like or YAP-dependent reprogramming of IECs occurs in various crypt injury models, these settings appear to involve a cell type conversion—that is, loss of stem cell and differentiation features and gain of fetal characteristics (Gregorieff et al., 2015; Nusse et al., 2018; Yui et al., 2018). In contrast, aVECs are differentiated with identifiable lineage properties, but they fail to fully mature into IECs and instead acquire a fetal-like program. Therefore, a delicate balance exists between cell maturation and repair in the setting of injury. We propose “adaptive differentiation” to describe how cell differentiation can be adaptive and meet novel needs in order to cope with adverse conditions. This paradigm may be conserved in other organs with high cell turnover rates.
Several mechanisms have been proposed for YAP’s regenerative function in the gut (Ayyaz et al., 2019; Gregorieff et al., 2015; Yui et al., 2018). As crypt-based cells are pliable to cell fate changes, depletion or overexpression of YAP induces profound cellular remodeling, including alteration of ISC identity, unintended lineage skewing, and loss or gain of proliferative activity. The action of YAP on other signaling pathways in the crypt likely explains a large portion of these effects. In the poly(I:C) injury model, we examined the function of YAP outside of the crypt niche. In doing so, we established a proliferation- and reprogramming-independent function of YAP during adaptive differentiation. In support of this, deletion of YAP did not block aVEC formation. Instead, YAP-deficient aVECs did not properly restore the villus barrier after injury. The two main markers of aVECs, the pro-invasion gene Msln and pro-survival gene Clu, may in fact be key functional targets of YAP necessary for repair (Chen et al., 2013; Trougakos et al., 2009). Thus, our data uncover a requirement for YAP in carefully tuning the function of aVECs. Interestingly, overexpression of YAP in ISCs leads to the formation of villi covered with epithelial cells that morphologically and transcriptionally resemble aVECs (Cheung et al., 2020), suggesting that YAP activation may be sufficient to trigger these unique repair cells.
An unresolved question stemming from our work is how and when YAP is induced following injury. Recent work using LGR5-DTR mice found minimal activation of YAP and fetal markers following ISC-specific ablation (Murata et al., 2020), indicating that stem cell loss alone and injury-induced dedifferentiation do not trigger YAP activation. To induce YAP activity, there is a threshold of damage that likely needs to be surpassed, coupled with an appropriate microenvironment (Romera-Hernandez et al., 2020; Taniguchi et al., 2015; Xu et al., 2020; Yui et al., 2018). Our study suggests that recovery from severe injury involves a transient intermediate step that is tailored to meet the demands of the repairing tissue prior to entering a pro-regenerative state. This has important implications for conditions such as Crohn’s disease, in which YAP is upregulated in the epithelium in ~60% of patients (Taniguchi et al., 2015; Yu et al., 2018). Furthermore, villus atrophy caused by small bowel injury is observed in a variety of enteropathies (Jansson-Knodell et al., 2018). Therapies designed around enhancing endogenous mechanisms of villus recovery (i.e., adaptive differentiation) may provide clinical benefit for patients with impaired healing capacity. While these adaptive responses appear to be beneficial for tissue repair, it will be important to determine whether maladaptive differentiation could potentially be a driver of pathology in settings of chronic inflammation and tumorigenesis.
Limitations of the study
Although our results support TA cells as the major source of aVECs, we cannot rule out the possible contribution of other intestinal cell types to the generation of aVECs, including CLU+ revSCs and pre-injury (homeostatic) villus cells. Though we found that BMI1+ cells did not expand after villus injury and contributed very little to aVEC formation, the relationship between BMI1+ and CLU+ cells is unknown. While the majority of villus cells are lost to poly(I:C)-induced injury, we were not able to confidently assess whether pre-injury villus cells can adapt to the aVEC state using Krt20CreER/R26RtdTomato mice given that both pre- and post-injury villus cells express KRT20. Creation of additional mouse strains encoding Cre drivers expressed only in the homeostatic intestine should further clarify the exact origins of aVECs in lineage tracing experiments. Finally, our study also highlights that YAP is a critical regulator of aVEC function, but the mechanism behind YAP’s actions and the relevant upstream inducers of YAP expressed in the lamina propria remain unclear. It will be important in future studies to establish what drives the aVEC state and what downstream targets of YAP are necessary for efficient repair.
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thaddeus S. Stappenbeck (stappet@ccf.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Sequencing data have been deposited at GEO under the accession codes GSE168439 (LCM-microarray) and GSE169718 (scRNA-seq).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
In vivo animal studies
C57BL/6J, Krt20-T2A-CreERT2, Lgr5-EGFP-IRES-CreERT2, Bmi1-CreER, Rosa-LSL-tdTomato, Vil1-Cre, Vil1-CreERT2, and Yapflox mice were obtained from the Jackson Laboratory. Yapflox mice were further backcrossed to the C57BL/6J strain for more than 4 generations. Experiments that called for only wild-type mice used 8-week-old C57BL/6J male mice. Pregnant C57BL/6J female mice were used to obtain embryos. All other experiments involving specific genetic strains used 7- to 10-week-old male and female mice with appropriate littermate controls. Mice were housed under specific-pathogen-free conditions and were maintained on a 12 h light/dark cycle. All animal studies were conducted in compliance with protocols approved by the Washington University Institutional Animal Care and Use Committee.
METHOD DETAILS
Animal procedures
For intraperitoneal injections, the following reagents were prepared and administered at the indicated dose. 1mg/mL poly(I:C) HMW (InvivoGen) was made according to the manufacturer’s instructions and 20mg/kg was injected. Tamoxifen (Sigma) was dissolved in corn oil to make 20mg/mL and 75mg/kg was injected. 10mg/mL CldU (Sigma) and 10mg/mL IdU (Sigma) dissolved in saline were injected at 42.5mg/kg and 57.5mg/kg, respectively. For intestinal permeability studies, mice were subjected to a 4 h fast during which food, water, and bedding were withdrawn from the cage. 4-kDa FITC-dextran (Sigma) was dissolved in PBS to make 100mg/mL, and 44mg/100g was delivered by oral gavage. 3 h after gavage, blood was collected by cardiac puncture, and serum was obtained in Microtainer tubes (BD). FITC-dextran levels were measured based on a standard curve on the Cytation 5 instrument (BioTek). For colonic biopsy injury, mucosal wounds were generated in the distal colon using biopsy forceps guided by a high-resolution miniaturized colonoscope as previously described (Seno et al., 2009).
Histology and immunostaining
Proximal small intestine (poly(I:C) injury) or distal colon (biopsy wounds) tissues were pinned out and fixed in 10% neutral buffered formalin overnight at 4°C. Fixed samples were washed in 70% ethanol three times and embedded in 2% agar (Sigma). This was followed by paraffin embedding, sectioning, and hematoxylin-eosin staining. For transmission electron microscopy, tissues were fixed in 2% paraformaldehyde/2.5% glutaraldehyde in 100mM cacodylate buffer and subsequently processed and imaged as previously described (Miyoshi et al., 2017).
Paraffin sections were de-paraffinized in xylene and rehydrated in isopropanol three times each. Antigen retrieval was performed in Trilogy solution (Sigma) for 20 min under boiling water. For immunohistochemistry (IHC), sections were additionally treated with 3% H2O2 in methanol to quench endogenous peroxidase activity. Slides were incubated in blocking solution (1% BSA/PBS containing 0.1% Triton X-100) for 1 h at room temperature before overnight treatment with primary antibodies diluted in blocking solution at 4°C. Slides were then treated with secondary antibodies diluted in blocking solution for 1 h at room temperature. For immunofluorescence (IF), sections were counterstained with Hoechst 33258 (Invitrogen) for 15 min and mounted in Fluoromount medium (Sigma). For IHC, sections were treated with VECTASTAIN Elite ABC-HRP Kit (Vector Laboratories), developed with DAB Peroxidase Substrate Kit (Vector Laboratories), counterstained with CAT hematoxylin (Biocare), and mounted in Cytoseal XYL solution (Thermo Fisher). Washes were performed in PBS.
RNAscope in situ hybridization
Intestinal tissues were pinned out and fixed in 4% paraformaldehyde overnight at 4°C. Fixed samples were incubated in 20% sucrose/PBS at 4°C for at least 8 h. Tissues were cryo-embedded in O.C.T. compound (Fisher Scientific) and sectioned at 7μm on a cryotome. In situ hybridization was performed on frozen sections using the RNAscope 2.5 HD Assay-RED Kit (ACDBio) according to the manufacturer’s instructions. Sections were mounted in EcoMount solution (Biocare). For co-staining with fluorescent antibodies, sections were treated with the appropriate antibodies using the IF protocol above following in situ hybridization staining.
Imaging and quantification
Bright-field images were acquired with an Olympus BX51 microscope. Fluorescent images were acquired with a Zeiss Axiovert 200M inverted microscope and a Zeiss Axio Imager M2 Plus wide field fluorescent microscope. Whole mount images were acquired with an Olympus SZX12 stereo dissection microscope. Live spheroid images were acquired with a Zeiss Cell Observer inverted microscope with color camera. The lengths of well-oriented villi/crypts/epithelial cells were measured using the cellSens software (Olympus). For histology-based quantifications, each data point represents an average value across 30–50 well-oriented villi/crypts in the proximal small intestine per animal (exact number is indicated in the figure legends). Nearest neighbor distances (NNDs) were quantified based on whole mount images using the NND plugin on Fiji (Schindelin et al., 2012). Images were processed with Adobe Photoshop CC.
LCM-microarray
Intestinal tissues were fixed in methacarn (60% methanol, 30% chloroform, 10% glacial acetic acid) for 4 h at room temperature and processed for paraffin embedding. Paraffin blocks were sectioned at 7μm on a microtome. Sections were de-paraffinized in xylene, rehydrated in a series of ethanol washes (70/95/100%), and briefly stained with methyl green (Vector Laboratories). Laser capture microdissection (LCM) was performed using CapSure LCM MicroCaps (Applied Biosystems) on the Arcuturus PixCell IIe system with an Olympus IX51 microscope base. RNA extraction and cDNA synthesis/amplification were carried out using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems) and Complete Whole Transcriptome Amplification Kit (WTA2, Sigma) according to the manufacturer’s instructions. Amplified cDNA products were purified with the QIAquick PCR Purification Kit (QIAGEN), and quality was assessed with the Agilent 2100 Bioanalyzer. Agilent mouse 8×60K v2 microarray chips (Agilent-074809) were used for hybridization. Microarray data from colonic WAE cells was previously reported and deposited (ArrayExpress E-MTAB-1175; Miyoshi et al., 2012). Data normalization, principal component analysis (PCA), and differential gene expression analysis were performed on the Partek software and visualized with Graphpad Prism 9. WAE and aVEC datasets were superimposed using Microsoft Excel and Prism 9. Gene ontology (GO) analysis and transcription factor motif enrichment of signature genes were pulled from ToppGene Suite (Chen et al., 2009) and Enrichr (Kuleshov et al., 2016). Gene set enrichment analysis (GSEA) of the Wnt signaling pathway (KEGG mmu04310), Crohn’s disease (Haberman et al., 2014), celiac disease (Dotsenko et al., 2021; Loberman-Nachum et al., 2019), fetal spheroid (Mustata et al., 2013) and YAP (Gregorieff et al., 2015) signatures (entire gene list or the top 300 genes in respective studies) were conducted using the GSEA v4 software (Subramanian et al., 2005). Normalized enrichment scores (NES) and false discovery rates (FDR q-value) are displayed.
ELISA
Intestinal tissues (<1 cm) were placed in Lysing Matrix D tubes (MP Biomedicals) containing RIPA buffer (Sigma) and protease/phosphatase inhibitors (Thermo Fisher) and lysed using the FastPrep-24 5G homogenizer (MP Biomedicals). Lysates were centrifuged for 10 min and supernatants were collected. Protein concentration was measured with the Pierce BCA Protein Assay Kit (Thermo Fisher). After normalization, PGE2 levels were measured using a Monoclonal PGE2 ELISA Kit (Cayman Chemical) according to the manufacturer’s instructions.
FACS
A standardized protocol for the isolation, dissociation, and sorting of IECs was previously described (Magness et al., 2013). The first 5 cm of the proximal small intestine was used for cell isolation. For downstream qRT-PCR analysis, poly(I:C)-damaged tissues from at least 2 mice were pooled together. Briefly, intestinal tissues were incubated in 30mM EDTA, first on ice for 20 min with 1.5mM DTT and second at 37°C for 10 min without DTT, and then shaken to lift the epithelium. After removal of the muscle layer, single cell dissociation was performed using an enzymatic cocktail containing 1mg/mL Collagenase/Dispase (Roche) and 0.2mg/mL DNase I (Roche) at 37°C for 10 min with intermittent shaking. The cell suspension was additionally subjected to vigorous pipetting and filtered through 70μm and 40μm strainers. Cells were washed in 10% FBS/PBS and resuspended in FACS buffer containing 2% BSA, 2mM EDTA, and 25mM HEPES in PBS. 10μM Y-27632 (ROCK inhibitor, R&D Systems) was added to every solution throughout the experiment. SYTOX-Red (Invitrogen) was used to assess cell viability. Flow cytometry and cell sorting were carried out on the Beckman Coulter MoFlo instrument.
qRT-PCR
Intestinal tissues (<5 mg) were homogenized in Buffer RLT Plus containing β-mercaptoethanol using the FastPrep system. 50,000 KRT20-tdT+ and 50,000 KRT20-tdT− cells from Krt20CreER/R26RtdTomato mice were sorted into Buffer RLT Plus containing β-mercaptoethanol. Total RNA was isolated using the RNeasy Plus Micro Kit (QIAGEN). cDNA was synthesized with iScript Reverse Transcription Supermix reagents (Bio-Rad). qRT-PCR was performed with iTaq Universal SYBR Green Supermix reagents (Bio-Rad) on a StepOne Real-Time PCR System (Applied Biosystems). Expression levels were normalized to B2m. Relative expression was calculated using the ΔΔCt method. Primers used in this study are listed in Table S3.
Single-cell spheroid growth assay
Generation of 50% L-WRN conditioned media for intestinal spheroid culture was previously described (Miyoshi and Stappenbeck, 2013). 10,000 KRT20-tdT+ and 10,000 KRT20-tdT− cells from Krt20CreER/R26RtdTomato mice were sorted into 50% L-WRN media, spun down, and resuspended in 40μL Matrigel (Corning). Cells were cultured in 50% L-WRN media containing 10μM ROCK inhibitor for 6 days. Media was changed once on day 3. Spheroid efficiency was calculated as the number of spheroids on day 6 over the number of seeded cells (10,000).
scRNA-seq and analysis
FACS purified IECs were resuspended in 10% FBS in DMEM/F12 (Sigma) for single-cell capture and sequencing. Single-cell libraries were constructed using Chromium Single Cell 3’ v3 reagents (10x Genomics) and sequenced on a NovaSeq6000 S4 system (Illumina) with around 50,000 reads per cell. Demultiplexing, alignment, and unique molecular identifier (UMI) counting were performed with Cell Ranger v4.0. For downstream analysis, filtered gene-barcode matrices generated by Cell Ranger were read into the Seurat package (v3.2.2) or Monocle 3 package (v2.3.0) on R (v4.0.2)/RStudio (v1.3.1056) (Butler et al., 2018; Cao et al., 2019; Trapnell et al., 2014). Single-cell transcriptomes from pediatric Crohn’s disease patients and healthy controls were obtained from the Gut Cell Survey (Elmentaite et al., 2020) and analyzed in Seurat. Plots generated from RStudio were adjusted with ggplot2 and processed with Adobe Illustrator CC.
In Seurat, low-quality cells with high (>9,000) and low (<500) unique gene counts and high mitochondrial counts (>20%) were first filtered out. The resulting data was log-normalized, and paired samples were integrated. PCA scores were computed on scaled data based on 2,000 of the most highly variable features. The first 15 PCs were used for graph-based clustering (resolution = 0.2–0.3), which was visualized with UMAP-based dimensional reduction (Becht et al., 2018). FindMarkers was used to identify cell types as well as to determine differentially expressed genes between Yapfl/fl and YapΔIEC cells. Immune cells (<1% of total cells) were removed from analysis. Gene expression levels were plotted with FeaturePlot, VlnPlot, and Dotplot functions. Enrichment of the fetal spheroid, YAP, and aVEC signatures were determined with the AddModuleScore function. GO analysis was performed with Enrichr.
In Monocle 3, the standard PCA method with 100 PCs was adopted to normalize the data and remove batch effects. Dimensionality reduction was conducted with UMAP, and cells were clustered based on community detection (resolution = 0.5). Cell types were identified with top_markers and plot_cells functions. After learning the trajectory graph (learn_graph), cells were ordered in pseudotime (order_cells) and the root node was selected according to where ISC markers (i.e., Olfm4) were most highly expressed. The plot_genes_in_pseudotime function (minimum expression = 0.5) was used to visualize gene expression as a function of pseudotime.
Statistics and reproducibility
Statistical analyses were performed in GraphPad Prism 9. P-values are indicated in the plots or figure legends with p < 0.05 denoted as significant. Data are expressed as mean ± standard deviation (SD). An unpaired two-tailed Student’s t-test was used when comparing two groups; a one-way ANOVA was used when comparing three or more groups; and a two-way ANOVA was used when comparing groups with two experimental variables. Data from independent experiments were pooled when possible. Otherwise, data are representative of at least two independent experiments. All images are representative of at least 3 animals. The investigators were not blinded during the study. Animals that had near 0% weight loss one day after poly(I:C) injection (<10% of mice) were excluded from the study as they did not exhibit intestinal damage. Further statistical details and quantification methods can be found in the figure legends.
Supplementary Material
Acknowledgements
We acknowledge the following Washington University in St. Louis facilities: DDRCC, GTAC, Siteman Cancer Center Flow Cytometry Core, and Molecular Microbiology Imaging Facility. Bill and Melinda Gates Foundation (OPP1098828, OPP1139330), Crohn’s & Colitis Foundation, National Institutes of Health (NIH), and Boehringer Ingelheim provided support. This work was funded by the following NIH grants: F30 DK120076 (T.E.O.); R01 DE025884, R01 AI134236, R01 DK124699, and R01 AI134035 (M.C.); R01 AT009741 and R01 DK122790 (T.S.S.).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Arvanitakis C (1979). Abnormalities of jejunal mucosal enzymes in ulcerative colitis and Crohn’s disease. Digestion 19, 259–266. [DOI] [PubMed] [Google Scholar]
- Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. [DOI] [PubMed] [Google Scholar]
- Bahar Halpern K., Massalha H, Zwick RK, Moor AE, Castillo-Azofeifa D, Rozenberg M, Farack L, Egozi A, Miller DR, Averbukh I, et al. (2020). Lgr5+ telocytes are a signaling source at the intestinal villus tip. Nat Commun 11, 1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Beumer J, and Clevers H (2021). Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol 22, 39–53. [DOI] [PubMed] [Google Scholar]
- Blanpain C, and Fuchs E (2014). Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, and Pan D (2010). The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev 24, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, et al. (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 15, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, and Brummelkamp TR (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17, 2054–2060. [DOI] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Hung WC, Wang P, Paul C, and Konstantopoulos K (2013). Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci Rep 3, 1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Xiol J, Dill MT, Yuan WC, Panero R, Roper J, Osorio FG, Maglic D, Li Q, Gurung B, et al. (2020). Regenerative Reprogramming of the Intestinal Stem Cell State via Hippo Signaling Suppresses Metastatic Colorectal Cancer. Cell Stem Cell 27, 590–604 e599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csillag C, Nielsen OH, Vainer B, Olsen J, Dieckgraefe BK, Hendel J, Vind I, Dupuy C, Nielsen FC, and Borup R (2007). Expression of the genes dual oxidase 2, lipocalin 2 and regenerating islet-derived 1 alpha in Crohn’s disease. Scand J Gastroenterol 42, 454–463. [DOI] [PubMed] [Google Scholar]
- de Sousa EMF, and de Sauvage FJ (2019). Cellular Plasticity in Intestinal Homeostasis and Disease. Cell Stem Cell 24, 54–64. [DOI] [PubMed] [Google Scholar]
- Di Sabatino A, Ciccocioppo R, Luinetti O, Ricevuti L, Morera R, Cifone MG, Solcia E, and Corazza GR (2003). Increased enterocyte apoptosis in inflamed areas of Crohn’s disease. Dis Colon Rectum 46, 1498–1507. [DOI] [PubMed] [Google Scholar]
- Dotsenko V, Oittinen M, Taavela J, Popp A, Peraaho M, Staff S, Sarin J, Leon F, Isola J, Maki M, et al. (2021). Genome-Wide Transcriptomic Analysis of Intestinal Mucosa in Celiac Disease Patients on a Gluten-Free Diet and Postgluten Challenge. Cell Mol Gastroenterol Hepatol 11, 13–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmentaite R, Ross ADB, Roberts K, James KR, Ortmann D, Gomes T, Nayak K, Tuck L, Pritchard S, Bayraktar OA, et al. (2020). Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev Cell 55, 771–783 e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Tumbar T, and Guasch G (2004). Socializing with the neighbors: stem cells and their niche. Cell 116, 769–778. [DOI] [PubMed] [Google Scholar]
- Gehart H, and Clevers H (2019). Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 16, 19–34. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, and Wrana JL (2015). Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718. [DOI] [PubMed] [Google Scholar]
- Guiu J, Hannezo E, Yui S, Demharter S, Ulyanchenko S, Maimets M, Jorgensen A, Perlman S, Lundvall L, Mamsen LS, et al. (2019). Tracing the origin of adult intestinal stem cells. Nature 570, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C, Buchen B, He GW, Hornef M, Torow N, Neumann H, Wittkopf N, Martini E, Basic M, Bleich A, et al. (2015). Caspase-8 controls the gut response to microbial challenges by Tnf-alpha-dependent and independent pathways. Gut 64, 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, Baldassano RN, Noe JD, Rosh J, Markowitz J, et al. (2014). Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 124, 3617–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BF, Ma Q, Henderson JW, Siddique A, Archer SY, and Hodin RA (2002). Enterocyte response to ischemia is dependent on differentiation state. J Gastrointest Surg 6, 403–409. [DOI] [PubMed] [Google Scholar]
- Holmes R, and Lobley RW (1989). Intestinal brush border revisited. Gut 30, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong AW, Meng Z, and Guan KL (2016). The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol 13, 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson-Knodell CL, Hujoel IA, Rubio-Tapia A, and Murray JA (2018). Not All That Flattens Villi Is Celiac Disease: A Review of Enteropathies. Mayo Clin Proc 93, 509–517. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R, and Arthur-Farraj P (2015). The Role of Cell Plasticity in Tissue Repair: Adaptive Cellular Reprogramming. Dev Cell 34, 613–620. [DOI] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent TH, and Moon HW (1973). The Comparative Pathogenesis of Some Enteric Diseases. Based on Cases Presented at the 22nd Annual Seminar of the American College of Veterinary Pathologists. Veterinary Pathology 10, 414–469. [Google Scholar]
- Kerzner B, Kelly MH, Gall DG, Butler DG, and Hamilton JR (1977). Transmissible gastroenteritis: sodium transport and the intestinal epithelium during the course of viral enteritis. Gastroenterology 72, 457–461. [PubMed] [Google Scholar]
- Kim HB, Kim M, Park YS, Park I, Kim T, Yang SY, Cho CJ, Hwang D, Jung JH, Markowitz SD, et al. (2017). Prostaglandin E2 Activates YAP and a Positive-Signaling Loop to Promote Colon Regeneration After Colitis but Also Carcinogenesis in Mice. Gastroenterology 152, 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Grompe M, and Sander M (2016). Stem cells versus plasticity in liver and pancreas regeneration. Nat Cell Biol 18, 238–245. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy ER (1988). Epithelial restitution in the gastrointestinal tract. J Clin Gastroenterol 10 Suppl 1, S72–77. [DOI] [PubMed] [Google Scholar]
- Lebwohl B, Murray JA, Rubio-Tapia A, Green PH, and Ludvigsson JF (2014). Predictors of persistent villous atrophy in coeliac disease: a population-based study. Aliment Pharmacol Ther 39, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loberman-Nachum N, Sosnovski K, Di Segni A, Efroni G, Braun T, BenShoshan M, Anafi L, Avivi C, Barshack I, Shouval DS, et al. (2019). Defining the Celiac Disease Transcriptome using Clinical Pathology Specimens Reveals Biologic Pathways and Supports Diagnosis. Sci Rep 9, 16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magness ST, Puthoff BJ, Crissey MA, Dunn J, Henning SJ, Houchen C, Kaddis JS, Kuo CJ, Li L, Lynch J, et al. (2013). A multicenter study to standardize reporting and analyses of fluorescence-activated cell-sorted murine intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 305, G542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manieri NA, Drylewicz MR, Miyoshi H, and Stappenbeck TS (2012). Igf2bp1 is required for full induction of Ptgs2 mRNA in colonic mesenchymal stem cells in mice. Gastroenterology 143, 110–121 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, Lakhdari O, Pineton de Chambrun G., Gareau MG, Broquet A, Lee GH, Shenouda S, Eckmann L, and Kagnoff MF (2013). TLR3, TRIF, and caspase 8 determine double-stranded RNA-induced epithelial cell death and survival in vivo. J Immunol 190, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Kljavin NM, Ybarra R, and de Sauvage FJ (2014). Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, and Stappenbeck TS (2012). Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, and Stappenbeck TS (2013). In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 8, 2471–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, VanDussen KL, Malvin NP, Ryu SH, Wang Y, Sonnek NM, Lai CW, and Stappenbeck TS (2017). Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J 36, 5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K., and Itzkovitz S (2018). Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis. Cell 175, 1156–1167 e1115. [DOI] [PubMed] [Google Scholar]
- Moore R, Carlson S, and Madara JL (1989). Villus contraction aids repair of intestinal epithelium after injury. Am J Physiol 257, G274–283. [DOI] [PubMed] [Google Scholar]
- Moss SF, Attia L, Scholes JV, Walters JR, and Holt PR (1996). Increased small intestinal apoptosis in coeliac disease. Gut 39, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Jadhav U, Madha S, van Es J, Dean J, Cavazza A, Wucherpfennig K, Michor F, Clevers H, and Shivdasani RA (2020). Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells. Cell Stem Cell 26, 377–390 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustata RC, Vasile G, Fernandez-Vallone V, Strollo S, Lefort A, Libert F, Monteyne D, Perez-Morga D, Vassart G, and Garcia MI (2013). Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep 5, 421–432. [DOI] [PubMed] [Google Scholar]
- Nardone G, Oliver-De La Cruz J., Vrbsky J, Martini C, Pribyl J, Skladal P, Pesl M, Caluori G, Pagliari S, Martino F, et al. (2017). YAP regulates cell mechanics by controlling focal adhesion assembly. Nat Commun 8, 15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto F, Klaus-Bergmann A, Ong YT, Alt S, Vion AC, Szymborska A, Carvalho JR, Hollfinger I, Bartels-Klein E, Franco CA, et al. (2018). YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M, Haelterman EO, and Burnstein T (1970). Transmissible gastroenteritis of swine: virus-intestinal cell interactions. I. Immunofluorescence, histopathology and virus production in the small intestine through the course of infection. Arch Gesamte Virusforsch 31, 321–334. [DOI] [PubMed] [Google Scholar]
- Ramig RF (2004). Pathogenesis of intestinal and systemic rotavirus infection. J Virol 78, 10213–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees WD, Tandun R, Yau E, Zachos NC, and Steiner TS (2020). Regenerative Intestinal Stem Cells Induced by Acute and Chronic Injury: The Saving Grace of the Epithelium? Front Cell Dev Biol 8, 583919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Hernandez M, Aparicio-Domingo P, Papazian N, Karrich JJ, Cornelissen F, Hoogenboezem RM, Samsom JN, and Cupedo T (2020). Yap1-Driven Intestinal Repair Is Controlled by Group 3 Innate Lymphoid Cells. Cell Rep 30, 37–45 e33. [DOI] [PubMed] [Google Scholar]
- Roulis M, Kaklamanos A, Schernthanner M, Bielecki P, Zhao J, Kaffe E, Frommelt LS, Qu R, Knapp MS, Henriques A, et al. (2020). Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature 580, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Tapia A, and Murray JA (2010). Classification and management of refractory coeliac disease. Gut 59, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffler MD, and Chaffee RG (1979). Small intestinal biopsy in a patient with Crohn’s disease of the duodenum. The spectrum of abnormal findings in the absence of granulomas. Gastroenterology 76, 1009–1014. [PubMed] [Google Scholar]
- Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, and Stappenbeck TS (2009). Efficient colonic mucosal wound repair requires Trem2 signaling. Proc Natl Acad Sci U S A 106, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers J, Zaalberg IC, and Maurice MM (2021). Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ 28, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, and Dignass AU (2008). Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroenterol 14, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svanes K, Ito S, Takeuchi K, and Silen W (1982). Restitution of the surface epithelium of the in vitro frog gastric mucosa after damage with hyperosmolar sodium chloride. Morphologic and physiologic characteristics. Gastroenterology 82, 1409–1426. [PubMed] [Google Scholar]
- Takeda H, and Kiyokawa E (2017). Activation of Erk in ileal epithelial cells engaged in ischemic-injury repair. Sci Rep 7, 16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagawa H, Takahashi I, Furuse M, Yoshitake-Kitano Y, Tsukita S, Ito T, Matsuda H, and Kiyono H (2003). Characteristics of claudin expression in follicle-associated epithelium of Peyer’s patches: preferential localization of claudin-4 at the apex of the dome region. Lab Invest 83, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, et al. (2015). A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trougakos IP, Lourda M, Antonelou MH, Kletsas D, Gorgoulis VG, Papassideri IS, Zou Y, Margaritis LH, Boothman DA, and Gonos ES (2009). Intracellular clusterin inhibits mitochondrial apoptosis by suppressing p53-activating stress signals and stabilizing the cytosolic Ku70-Bax protein complex. Clin Cancer Res 15, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDussen KL, Stojmirovic A, Li K, Liu TC, Kimes PK, Muegge BD, Simpson KF, Ciorba MA, Perrigoue JG, Friedman JR, et al. (2018). Abnormal Small Intestinal Epithelial Microvilli in Patients With Crohn’s Disease. Gastroenterology 155, 815–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chiang IL, Ohara TE, Fujii S, Cheng J, Muegge BD, Ver Heul A, Han ND, Lu Q, Xiong S, et al. (2019). Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells. Cell 179, 1144–1159 e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tang Y, Sheng X, Tian Y, Deng M, Du S, Lv C, Li G, Pan Y, Song Y, et al. (2020). Secreted stromal protein ISLR promotes intestinal regeneration by suppressing epithelial Hippo signaling. EMBO J 39, e103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Luo Y, Cong Z, Mu Y, Qiu Y, and Zhong M (2018). MicroRNA-590–5p Inhibits Intestinal Inflammation by Targeting YAP. J Crohns Colitis 12, 993–1004. [DOI] [PubMed] [Google Scholar]
- Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, et al. (2018). YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 22, 35–49 e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, and Piccolo S (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, et al. (2020). TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Wei H, Sun R, and Tian Z (2007). Recognition of double-stranded RNA by TLR3 induces severe small intestinal injury in mice. J Immunol 178, 4548–4556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited at GEO under the accession codes GSE168439 (LCM-microarray) and GSE169718 (scRNA-seq).