Figure 6. The PAR-3 size threshold in an endogenous setting.
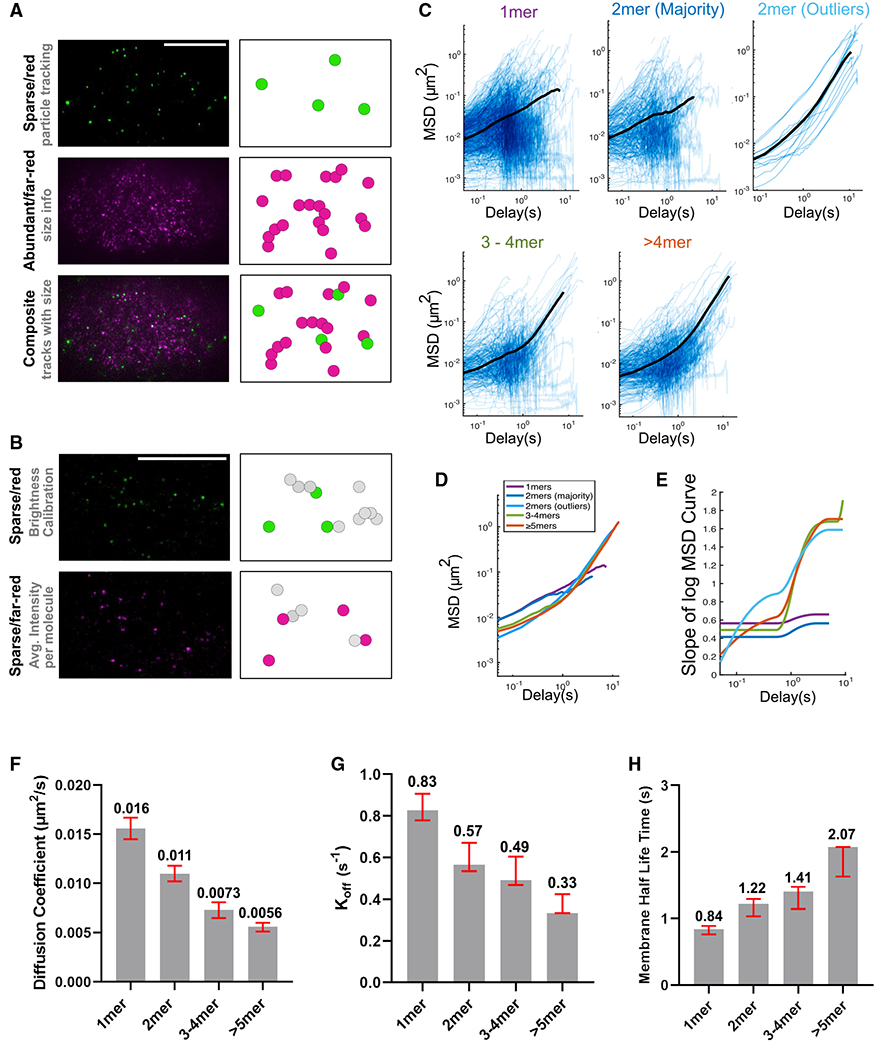
(A) TIRF images and cartoon illustrations of the dual-labeling experiment. Magenta represents far-red/abundant channel and green represents red/sparse channel. Scale bar represents 10 μm.
(B) TIRF images and cartoon illustrations of the calibration double dilution experiment, where the dyes for both channels are diluted to single-molecule levels. Scale bar represents 10 μm.
(C) Log/log scale MSD curves for each cluster group binned by estimated size, each curve describing the motion of a single PAR-3 cluster. For each plot, data were acquired and pooled from five embryos. Averaged MSD curves are shown for time lags defined by at least 20 data points except for the dimer outlier curves (top right), for which the average is over all 13 particles observed.
(D) Comparison of the averaged MSD curves for each cluster group binned by estimated size.
(E) Slopes of the averaged MSD curves for each cluster group binned by estimated size. Slopes were estimated by fitting a smoothing spline to each curve and then taking its derivative.
(F) The diffusion coefficient measured in a dual-labeling experiment. Red bars indicated 95% CI. Data points were pooled from three embryos for each strain. n = 672, 297, 244, and 322 particles for monomers, dimers, trimers to tetramers, and greater than pentamers, respectively.
(G) The koff measured in a dual-labeling experiment. Red bars indicated 95% CI. Data points were pooled from three embryos for each strain. n = 672, 297, 244, and 322 particles for monomers, dimers, trimers to tetramers, and greater than pentamers, respectively.
(H) The membrane binding lifetime measured in a dual-labeling experiment. Red bars indicated 95% CI. Data points were pooled from three embryos for each strain. n = 672, 297, 244, and 322 particles for monomers, dimers, trimers to tetramers, and greater than pentamers, respectively.
