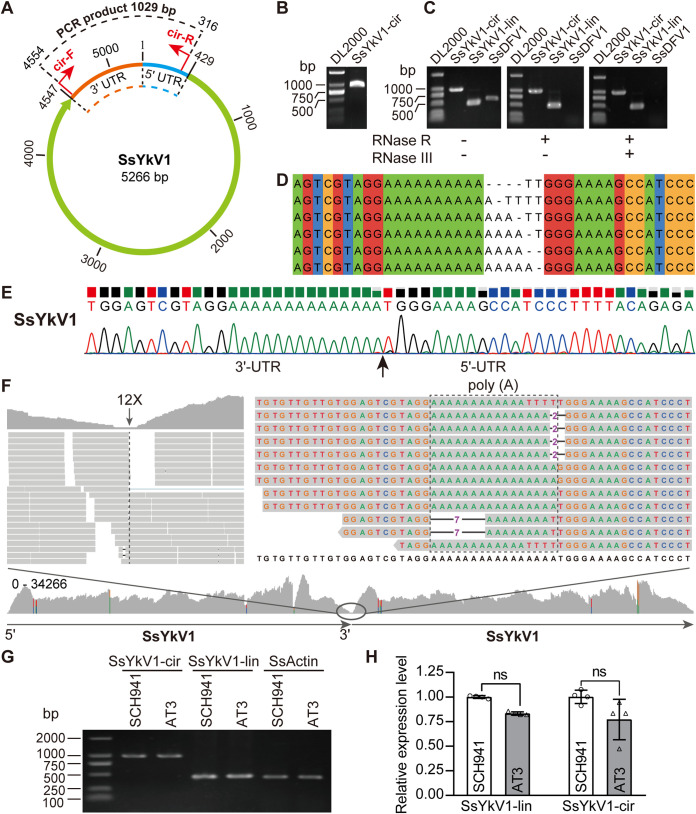
FIG 6.
Identification of the circular genomic form of SsYkV1. (A) Schematic diagram of the circular genomic form of SsYkV1. The positions of the divergent primers used to verify the circular genome are marked with red arrows. (B) Inverse RT-PCR validation of the circular genome using divergent primers. The total RNAs of strain SCH941 were used as the template for the reverse transcription reactions. (C) The circular (cir) genome of SsYkV1 was detected by PCR using divergent primers. RNase R and RNase III-treated or untreated viral RNAs of strain SCH941 were used as the template for reverse transcription reactions. lin, linear cir, circular. (D) Sanger sequencing showed the variability in the number of adenine (A) residues at the 3′ terminus and thymine (T) at the 5′ terminus of SsYkV1. (E) Sanger sequencing chromatogram of inverse RT-PCR products confirmed the head-to-tail site of the SsYkV1 circular genome. (F) Mapping of NGS reads from strain SCH941 onto the circular genome of SsYkV1. The template for mapping contains two genomes connected head to tail. The genomic sequence variation in the junction region is enclosed by a dashed box. (G) Semiquantitative RT-PCR analysis of the circular RNA levels of SsYkV1 in strains SCH941 and AT3. Thirty cycles were used for semiquantitative RT-PCR. (H) RT-qPCR was performed to verify the circular RNA levels of SsYkV1. ns, not significant.