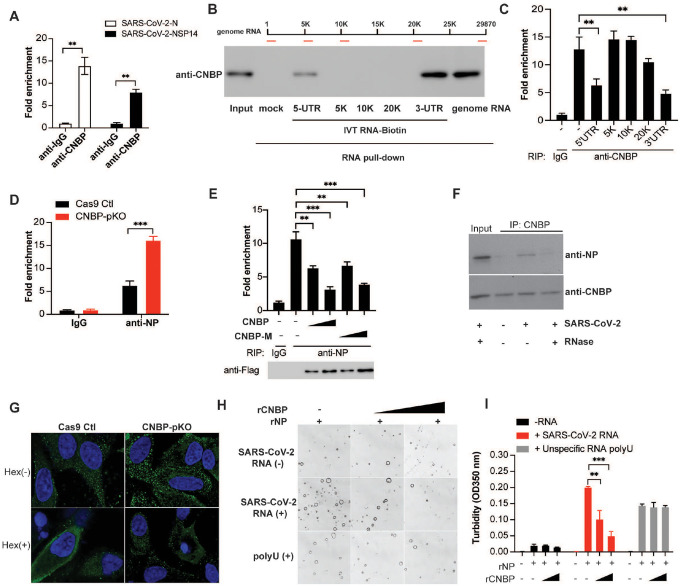
Figure 3. CNBP binds viral RNA competing with NP leading to disruption of viral RNA-nucleocapsid protein condensates.
(A) RIP assay with hACE2-A549 cell lysates prepared after 24 h of infection with SARS-CoV-2 by using anti-CNBP or control immunoglobulin. Immunoprecipitated SARS-CoV-2 positive-strand RNA was quantified by RT-qPCR. (B) RNA pull-down assay showing the binding activity of SARS-CoV-2 RNA genome or in vitro-transcribed (IVT) RNAs to CNBP. (C) RIP assay and RT-qPCR analysis of the binding activity of CNBP with SARS-CoV-2 genome RNA in the present of the indicated IVT RNAs. (D) RIP assay with A549 WT or CNBP pKO cell lysates prepared after 24h of infection with SARS-CoV-2 by using anti-NP. The immunoprecipitated SARS-CoV-2 positive-strand RNA was quantified by RT-qPCR. (E) CNBP pKO transfected with CNBP and CNBP-M, cell lysates were prepared after 24h of infection with SARS-CoV-2, the interaction of SARS-CoV-2 positive-strand RNA with NP was analyzed by RIP assay and RT–qPCR analysis as described in D. (F) Co-immunoprecipitation of CNBP and NP protein in SARS-CoV-2-infected cell lysates treated with or without RNase. (G) Increased NP puncta are formed in CNBP pKO cells compared with Cas9 Ctl hACE2-A549 cells infected with SARS-CoV-2 and disrupted by treating cells with 1,6-hexanediol. (H) NP protein LLPS were observed under bright field of a confocal microscope and could be disrupted by the addition of rCNBP. (I) The turbidity of each sample was measured by absorbance at 350 nm.