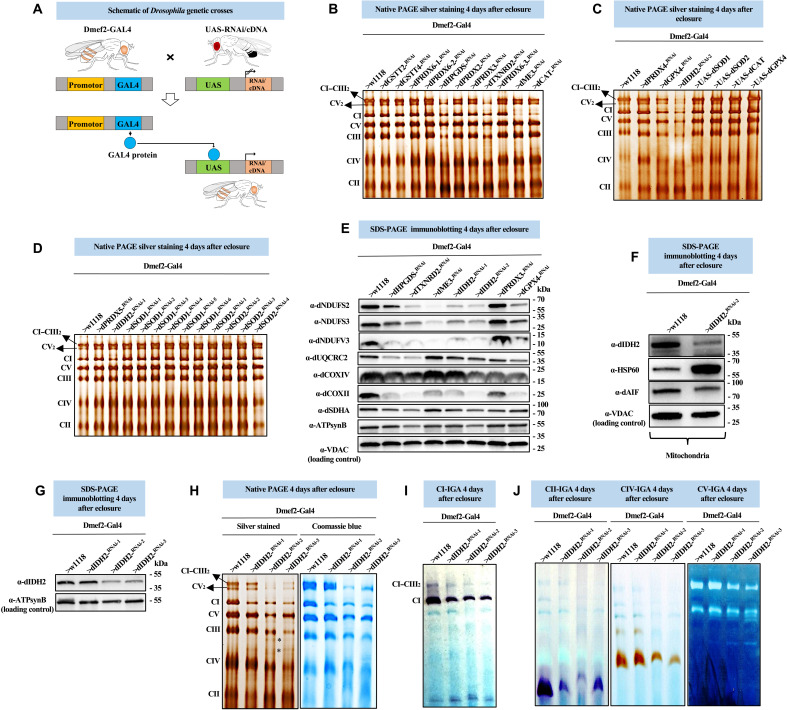
Fig. 1. RNAi-mediated disruption of the Drosophila ortholog of IDH2 (CG7176) and other antioxidant enzymes in Drosophila flight muscles impairs OXPHOS assembly.
(A) Schematic describing the Gal4-UAS system. (B to D) Mitochondrial preparations from adult flight muscles expressing the UAS-RNAi or UAS-cDNA constructs shown in muscles using Dmef2-Gal4 were analyzed by silver staining of native gels (see Materials and Methods). Samples expressing RNAi constructs for dME3 were analyzed 2 days after eclosure due to early lethality. For some of the dSOD1 and dSOD2 RNAi lines, lethality began by 3 days, but a few flies survived to 5 days. Hence, we dissected flies at 4 days. (E) Total tissue lysates from flight/thoracic muscles isolated from flies with the genotypes listed were analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti-dNDUFS2, anti-NDUFS3 (which detects dNDUFS3), anti-dNDUFV3, anti-dUQCRC2, anti-dCOXIV, anti-dCOXII, and anti-dSDHA. (F) Mitochondrial preparations from thoraces of flies with the genotypes listed were analyzed by SDS-PAGE and immunoblotting with anti-dIDH2, anti-dAIF, and anti-Hsp60 (which detects dHsp60). (G) Total tissue lysates from flight/thoracic muscles from flies with the genotypes shown were analyzed by SDS-PAGE and immunoblotting with anti-dIDH2. (H) Mitochondrial preparations from thoraces of flies with the genotypes listed were analyzed by BN-PAGE followed by silver staining (left panel) and Coomassie blue staining (right panel). The asterisk (*) denotes accumulated assembly intermediates (AIs). (I and J) CI, CII, CIV, and CV in-gel activity (IGA) assays. See figs. S1 to S3 for replicates and quantification of immunoblots in (E) to (G). Loading controls used in the western blots are as indicated.