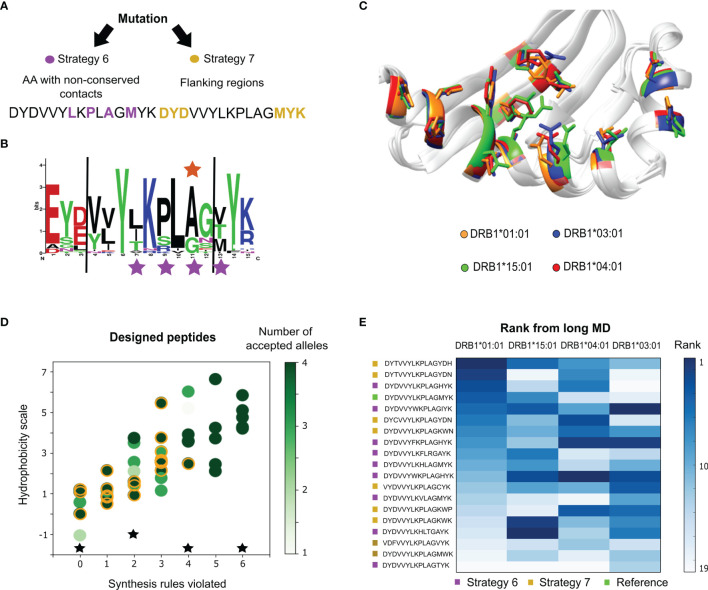
Figure 3.
P. vivax epitope design for multiple-allele binding. (A) Mutation strategies implemented by randomly modifying amino acids in the core region with low conservation (strategy 6), and those belonging to the peptide flanking regions (strategy 7). (B) Logo representation of the alignment showing the frequency of the epitope amino acids at each position. The positions modified during design strategy 1 are marked with purple stars, and the reported polymorphism is marked with an orange star. (C) Structural view of the polymorphic amino acids from the MHC II β-chain. (D) Hydrophobicity values, the number of synthesis rules violated and the number of alleles where the consensus score criterion was improved for the designed peptides. From a total of 42 peptides, 18 were selected for long MD simulations (orange circles). The controls are represented as black stars. (E) Average rank of the peptide candidates for the four alleles estimated from the long MD. The darker the shade of blue, the higher the rank. The peptides are color-coded according to the two mutation strategies from panel (A) The reference peptide is labeled green.