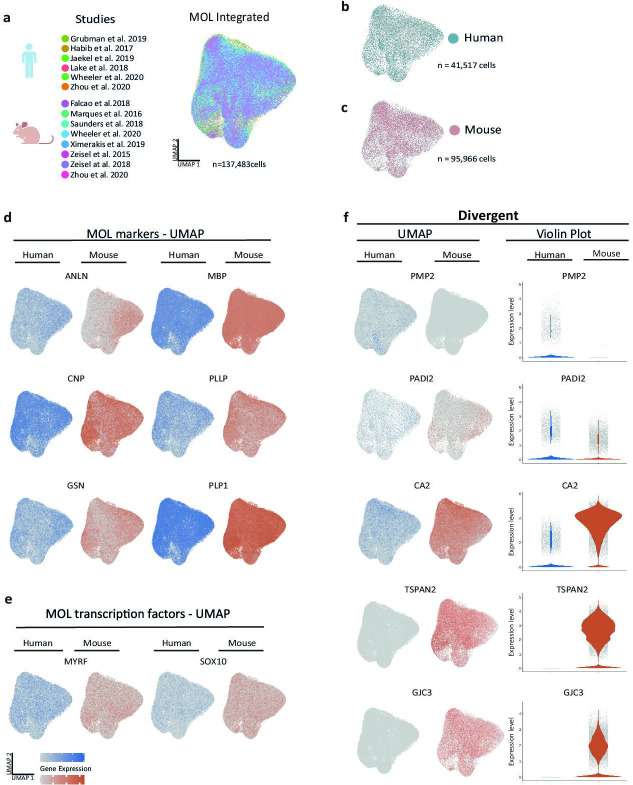
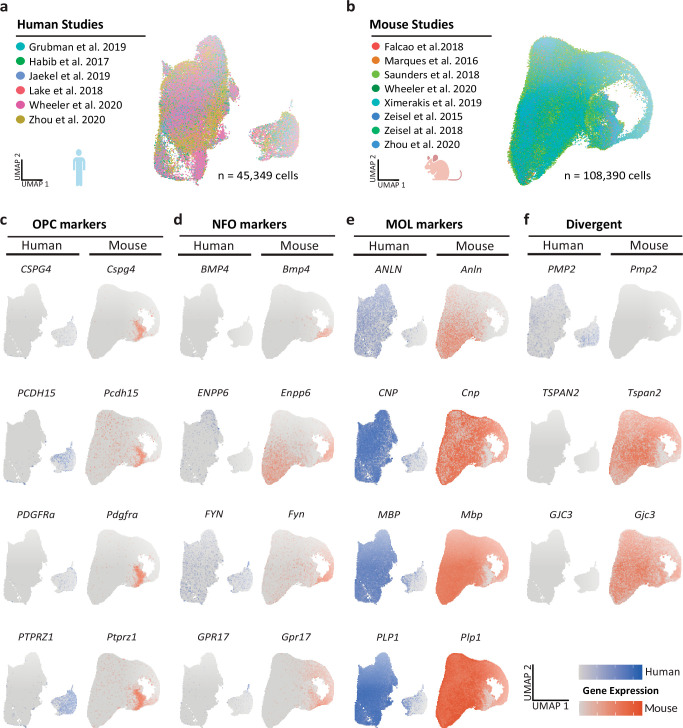
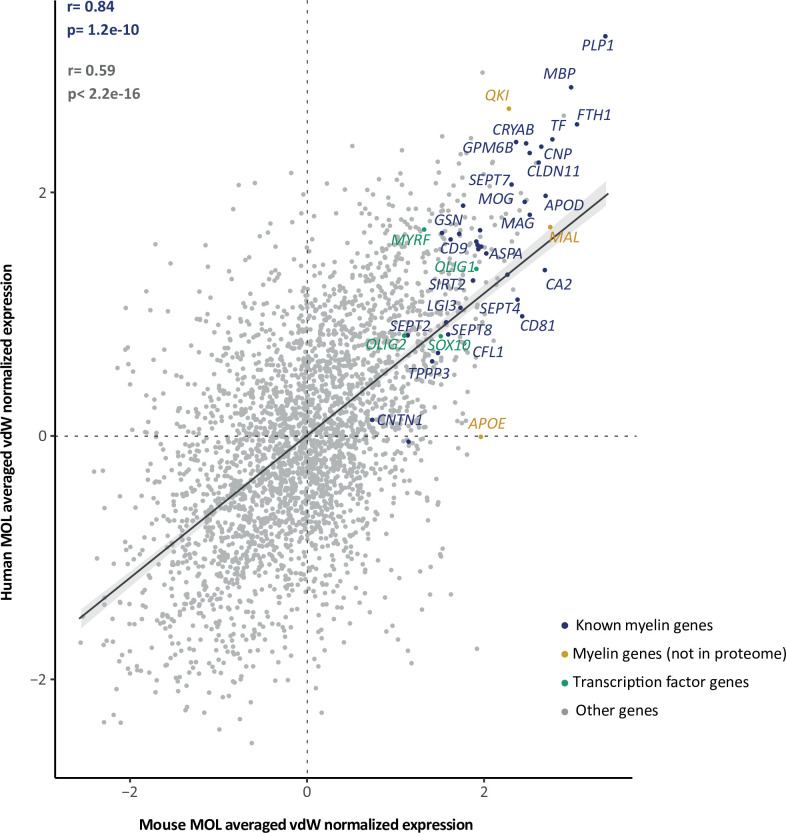
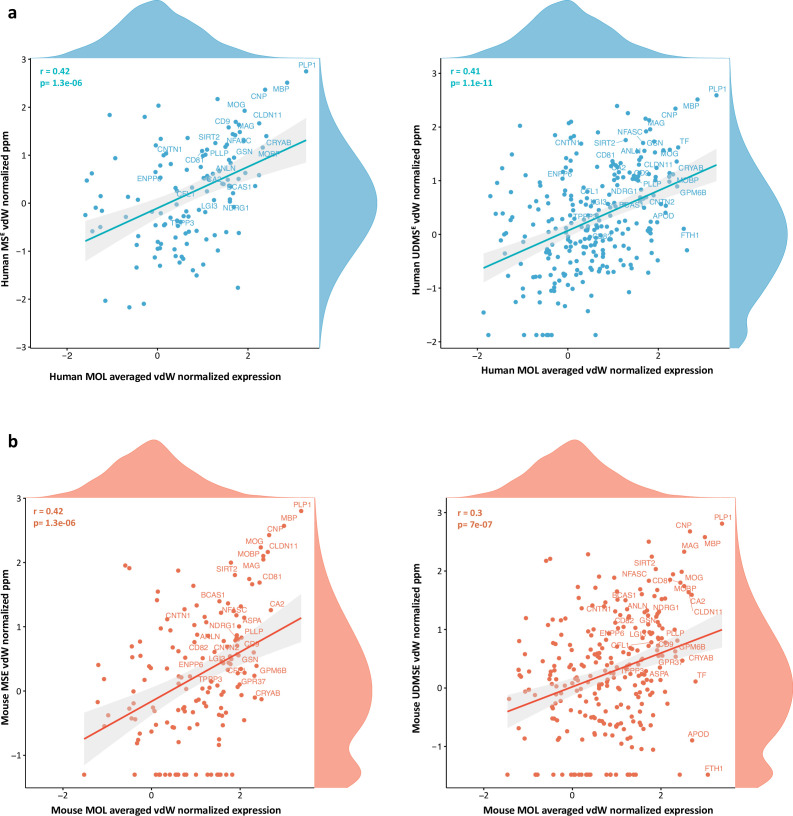
Figure 3. Cross-species scRNA-seq profile comparison of mature oligodendrocytes (MOL).
(a–c) Uniform manifold approximation and projection (UMAP) plot of the scRNA-seq profile of MOL integrated from previously established human (b) and mouse (c) datasets. In (a), cells contributed by distinct studies are highlighted in different colors; the corresponding references are given. (d, e) UMAP feature plots highlighting expression of selected MOL marker genes (d) and transcription factors (e) in the integrated object comprising MOL of both humans (blue) and mice (orange). (f) UMAP feature plots and violin plots exemplify genes that display preferential expression in MOL of humans (PMP2, PADI2) or mice (GJC3, TSPAN2, CA2).