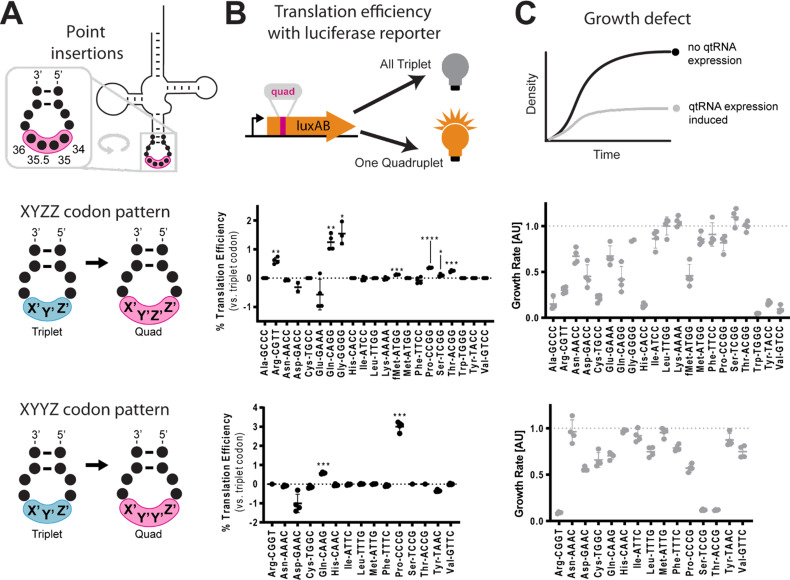
Figure 2. Engineering qtRNAs with codon patterns.
(A) We measured quadruplet tRNAs (qtRNAs) that might arise through point insertions in the anticodon loop.
Each qtRNA is based upon a tRNA from the Escherichia coli genome that serves as a ‘scaffold’ (Supplementary file 1). We tested two quadruplet codon patterns: a tRNA decoding the triplet codon ‘XYZ’ to a qtRNA decoding the quadruplet codon ‘XYZZ’ or ‘XYYZ’. In instances in which XYZZ and XYYZ are the same, the qtRNA is depicted on the XYZZ graph. We use ‘qtRNA’-‘three letter scaffold’-‘four letter codon’ nomenclature to refer to qtRNAs; for example, a serine qtRNA bearing a 5’-UCUA-3’ anticodon that recognizes 5’-UAGA-3’ in mRNA transcripts is referred to as qtRNASerTAGA. (B) We measured qtRNAs using a luciferase readthrough assay. Measurements are taken kinetically and normalized to culture density, and efficiency is reported relative to luminescence produced by a wildtype (WT), all triplet luciferase transcript. qtRNAs that are statistically >0 are annotated with their one-sample t-test p-value: 0.033(*), 0.0021(**), 0.0002(***), < 0.001(****). fMet qtRNAs are measured with a luciferase reporter bearing a quadruplet codon at residue 1; all others are measured with a quadruplet codon at residue 357 of luxAB. (C) Expression of qtRNAs can be toxic. Here, we report the fractional OD600 density difference between cultures where qtRNA expression had been induced versus suppressed. Data in (B and C) represent the mean and standard deviation of three to eight technical replicates in one biological replicate. For raw data, see Figure 2—source data 1.