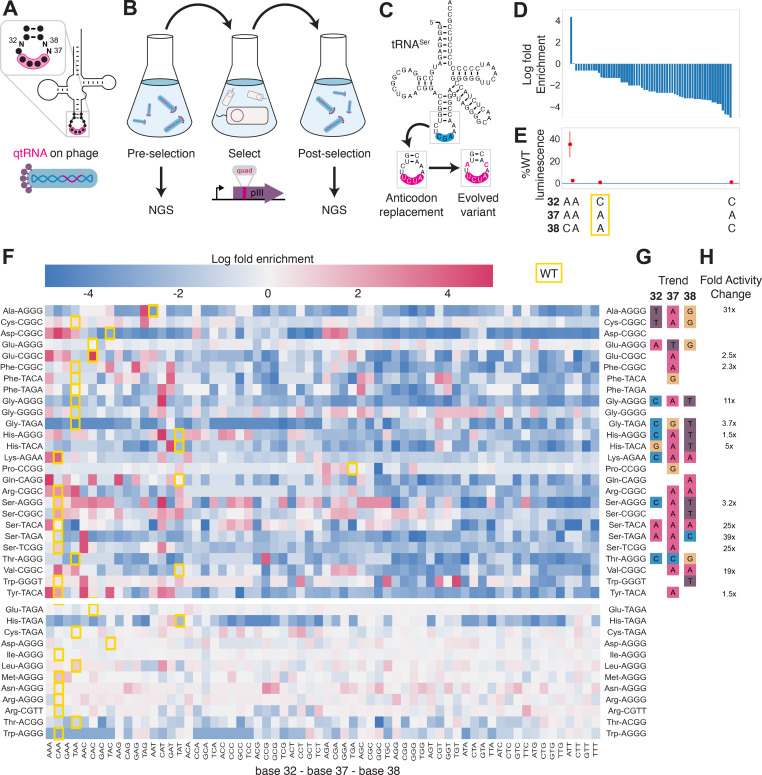
Figure 4. Directed evolution of anticodon loop sides.
(A) We created quadruplet tRNAs (qtRNA) libraries using degenerate primers to randomize positions 32, 37, and 38 of the anticodon. The qtRNA library is expressed from a ΔpIII M13 bacteriophage. (B) We selected these libraries by challenging the phage to infect and propagate in bacteria that require a quadruplet codon to be translated in order to produce a functional version of the essential phage gene, pIII. Libraries were next-generation sequencing (NGS) sequenced to >10× library size before and after selection. (C) tRNASerTCG is known to be a scaffold for the functional qtRNASerTAGA after anticodon replacement alone. Additional mutations to the sides of the anticodon loop are known to improve quadruplet codon translation efficiency. (D) Log fold enrichment of the population abundance and (E) translation efficiency as measured by a luciferase readthrough assay of the 64 possible combinations of nucleotides at positions 32, 37, and 38. (F) Log fold enrichment of all 41 qtRNA libraries for each of the 64 library members. Libraries are separated by those that exhibit abundance changes during selection (above) from those that exhibit no significant abundance changes (below). The anticodon loop sides present in the wildtype (WT) tRNA scaffold are boxed in gold. For raw data, see Figure 4—source data 1. (G) For each library, the trend in nucleotide preference for each position is listed. (H) Nucleotide preferences for select libraries were measured by cloning a qtRNA variant and measuring it using a luciferase readthrough assay. The fold improvement in activity over the WT values of base 32, 37, and 38 are listed. In (e and f), the original identities of bases 32, 37, and 38 found in the WT triplet tRNA scaffold are boxed in gold.