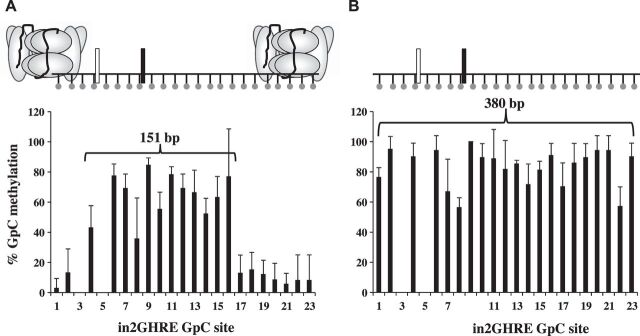
Fig. 3.
Nucleosome position analysis by nucleosome occupancy and methylome sequencing (NOMe-seq) across the IGF-1 in2GHRE from nuclei and purified genomic DNA of 6 wk old mouse livers. The %GpC methylation results following M.CviP1 treatment and indicated presence or absence of nucleosomes on in2GHRE for nuclei of 6 wk old mice (n = 4) showing a nucleosome-depleted region of at least 151 bp (A) compared with purified liver genomic DNA of 6 wk old mice (n = 3) (B) showing all sites (380 bp) are accessible. Each GpC site is represented by a ball and stick. Results for GpC sites 3 and 5 were in the context of GCG and therefore excluded due to interpretation inability. Relative locations of the 2 STAT5b binding sites are represented by white and black rectangles.