Table 3.
Comparison of different blockers for envelope protein (E) of SARS-CoV-2, blockers' structures, and software used for docking.
| Compound | Chemical structure | Target | Software | Ref. |
|---|---|---|---|---|
| Mefenamic acid | 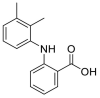 |
E protein | Docking server CB-Dock, Automated cavity-detection, AutoDock Vina, MD by GROMACS | [56] |
| Artemether | 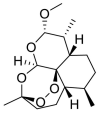 |
E protein | Docking server CB-Dock, Automated cavity-detection, AutoDock Vina, MD by GROMACS | [56] |
| Gliclazide | 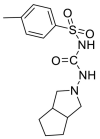 |
E protein | Experimental test | [71] |
| Memantine | 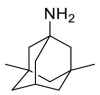 |
E protein | Experimental test | [71] |
| Wortmannin | 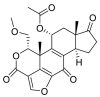 |
E protein | HDOCK web(PPD), Swiss dock web-interface, MD by Nanoscale molecular dynamics (NAMD) | [72] |
| Veliparib | 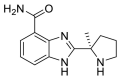 |
E protein | Schrodinger Glide software in SP mode | [73] |
| Rimantadine | 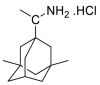 |
E protein | DOCK6 program, MD by GROMOS | [74,75] |
| Nimbolin | 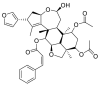 |
E protein | AutoDock Vina, MD by GROMACS | [76] |
| 7-Deacetyl-7-benzoylgedunin | 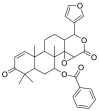 |
E protein | AutoDock Vina, MD by GROMACS | [76] |
| 24-Methylenecycloartanol | 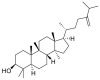 |
E protein | AutoDock Vina, MD by GROMACS | [76] |
| Plerixafor | 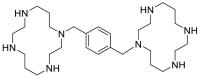 |
E protein | – | [77] |
| Mebrofenin | 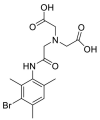 |
E protein | – | [77] |
| Kasugamycin (hydrochloride hydrate) | 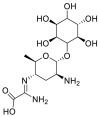 |
E protein | – | [77] |
| Saroglitazar magnesium | 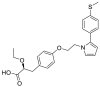 |
E protein | – | [77] |
