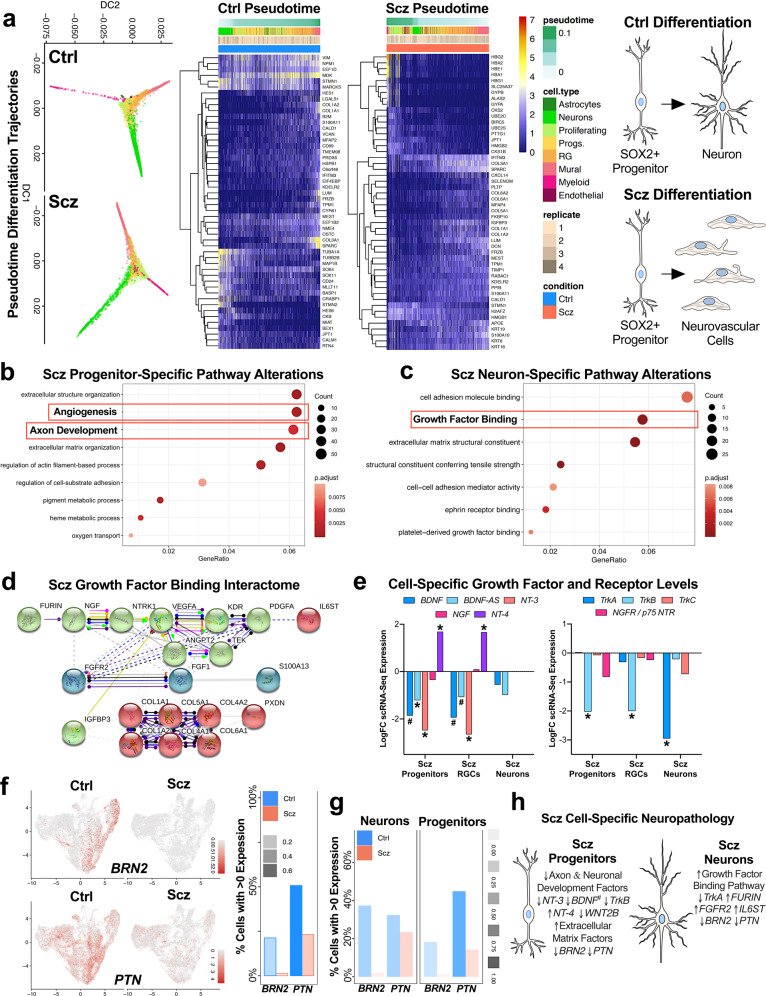
Fig. 3. Altered cell-lineages and cell-specific neuropathology in Scz organoids.
a Computational analyses reveal altered single-cell lineages in Scz organoids. Diffusion maps of single-cell differentiation trajectories were generated for all 26,335 cells via harmonized principal components. This revealed similar, but subtly altered, differentiation trajectories in Scz organoids (a). Heat-maps of slingshot-identified genes most-associated with pseudotime cell lineages of Scz organoids are also presented. Note differences in enrichment for collagen and matrix organization factors in Scz single-cell transcriptomes, whereas Ctrl organoids tended to exhibit up-regulation of neuronal factors (e.g., STMN2, TUBB2B, CRABP1) as expected. Notably, markers such as IFITM3 and POU5F1 (OCT4), as well as cell adhesion and vascularization factors, segregated Scz from Ctrl transcriptomes within pseudotime trajectories. Expression of these markers did not appear to identify a broader pool of undifferentiated stem cells, but rather indicated a series of intrinsic alterations within pseudotime trajectories of Scz cell-types and their differentiation patterns. These pseudotime differentiation trajectory analyses broadly replicated data presented in Figs. 1–2, notably disrupted neuronal differentiation dynamics that contributed to fewer total neurons in Scz organoids. In addition, these computational data support the notion that progenitors are diverted towards altered lineages (notably, neuroendothelial and vascularization-related lineages, see Fig. 2f) via differentially enriched gene-sets in Scz organoids. The net result of these transcriptional differences is that while Scz progenitors and organoids produce some neurons, vascular-related cell-types tend to be overproduced in Scz organoids. b Scz progenitors exhibit disrupted transcription of axon development factors. SOX2+ progenitor scRNA-Seq transcriptomes were isolated and differentially expressed genes (DEG) examined (5% FDR threshold average logFC Scz/Ctrl ≤2.5, and >3). This revealed down-regulation of a gene-set involved in axon development. Other DEG targets in Scz progenitors by biological process are presented here as a dotplot, and resulted in gene-set enrichment for extracellular organization, adhesion, as well as angiogenesis in Scz progenitors. These differentially enriched gene-sets further support the notion that Scz progenitors exhibit cell-specific transcriptional remodeling, which is consistent with the fact that very early progenitor cell-types retain the ability to putatively differentiate into neuronal or neuroendothelial lineages. c–d, Scz neurons exhibit pathway enrichment for growth factor binding. Similar to progenitors, neuron scRNA-Seq transcriptomes were examined for DEGs (5% FDR threshold average LogFC Scz/Ctrl ≤ 2.5, and >3). Analysis of gene ontologies by molecular function revealed enrichment for “growth factor binding” in Scz neurons (c), which was defined by down-regulation of TrkA (NTRK1) neurotrophin receptor gene expression and up-regulation of factors such as FURIN (neurotrophin pro→mature processing), FGFR2, PDGFA, TGFBR3, and the interleukin-6 signal transducer IL6ST (d). Protein–protein interactions between enriched growth factor binding factors in Scz neurons revealed notable interplay and interaction pathways (e.g., Furin →NGF→NTRK1) that place neuron-specific DEGs such as IL6ST and S100A13 as downstream targets of growth factor binding enrichment (d). Alternatively, these downstream factors may be putatively repressed by the inhibitory interaction effect of IGFBP3 on VEGFA (d), which are prototypic markers of neurovascular cells which also exhibited increased abundance within Scz organoids (see Fig. 2f). Other pathway analysis (e.g., KEGG) identified altered Pl3K-Akt signaling in Scz neurons (see Fig. S4). e Cell-specific disruption of neurotrophic factors and receptors in Scz organoids. Given unbiased detection of alterations in NT-3 (NTF3) in Scz progenitors and TrkA (NTRK1) receptors in Scz neurons, we next examined progenitor cell-types for specific differences in neurotrophins (BDNF, BDNF-Antisense/BDNF-AS, NGF, NT-3, and NT-4; left) and their cognate tyrosine kinase receptors (TrkA, TrkB, TrkC, & p75 NTR/NGFR; right). SOX2+ progenitors and radial glial cell (RGC) progenitors examined almost identical neurotrophin dysregulation and were defined by the trend for decreased expression of BDNF, BDNF-AS, NT-3, and TrkB expression. Curiously, NT-4 was up-regulated in Scz progenitor cell-types. Scz neurons were defined only by their near 3-fold reduction of TrkA gene expression. Thus, Scz progenitors and neurons exhibit a developmental “switch” in neurotrophic growth factor pathology within organoids. f–g, Validating gene-expression differences of candidate proteome-derived targets, BRN2 (POU3F2) and PTN, in specific cell-types of Scz organoids. Candidate proteome targets (Fig. 2e) were cross-examined for differences at the single-cell level (see Fig. S5 for expression of proteome targets in global, progenitor-only and neuron-only scRNA-Seq datasets). Given survival, differentiation, and diminished growth factor support, here we emphasize the neuronal transcription factor BRN2 (POU3F2) and putative growth factor PTN as putative targets for mechanism experiments. Scz progenitors exhibited almost complete depletion of BRN2 relative to Ctrl progenitors, indicating disrupted induction of neuronal differentiation. PTN expression was disrupted in both Scz progenitors and neurons but exhibited a greater difference in progenitors. h Resolving cell-type specificity of Scz neuropathology in 3D cerebral organoids. Schematic summary of cell-specific alterations in Scz progenitors (left) and neurons (right). Scz progenitors exhibited entropy in neurotrophic growth factors, specific reduction of TrkB (NTRK2), and depletion of axon development factors. Scz progenitors also exhibit enrichment for extracellular structure, matrix organization, and angiogenesis factors, which partially explains the intrinsic alteration in cell lineage (Fig. 3a) and differentiation trajectories towards neuroendothelial factors and cell types (Figs. 2, 3). Total scRNA-Seq dataset comprised n = 26, 335 cells, n = 20,844 genes from 7 iPSC lines; Ctrl n = 15,089 scRNA-Seq transcriptomes from 4 Ctrl iPSC lines, and Scz n = 11, 246 scRNA-seq transcriptomes from 3 Scz iPSC lines. * significant p and FDR values. # = p value significant, with a final FDR score at the cut-off threshold. Ctrl control, Scz schizophrenia.