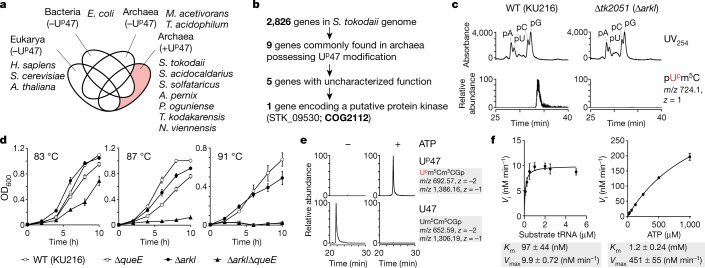
Fig. 3. Identification and characterization of the RNA kinase responsible for Up47 and its physiological role.
a, Venn diagram showing unique and shared genes among the Bacteria (E. coli), Eukarya (Homo sapiens, S. cerevisiae and Arabidopsis thaliana) and Archaea (Methanosarcina acetivorans, Thermoplasma acidophilum, S. tokodaii, Sulfolobus acidocaldarius, Saccharolobus solfataricus, Aeropyrum pernix, Pyrobaculum oguniense, T. kodakarensis and Nitrososphaera viennensis) domains possessing (+) or lacking (–) Up47. The pale red area includes genes unique to archaea having Up47. b, Comparative genomic analysis performed to narrow down the candidate genes responsible for Up47 modification. c, LC–MS nucleotide analysis of tRNA fractions from wild-type (WT, KU216) (left) and Δtk2051 (right) strains of T. kodakarensis. The upper panel shows the UV trace at 254 nm. The peaks for pA, pU, pC and pG are marked. The lower panel shows the XIC for the proton adduct of the dimer pUpm5C (m/z 724.1, z = 1). d, Growth measurement (OD600) of wild-type (KU216) (open circles), ΔqueE (squares), ΔarkI (closed circles) and ΔarkIΔqueE (triangles) strains of T. kodakarensis at 83 °C (left), 87 °C (middle) and 91 °C (right). Data represent the average values of technical triplicates ± s.d. e, In vitro reconstitution of Up47 with recombinant TkArkI in the presence (right panels) or absence (left panels) of ATP. XICs show the sum of monovalent and divalent negative ions from RNase T1-digested fragments containing Up47 (upper panels) or U47 (lower panels). f, Kinetic measurements of in vitro Up47 formation by TkArkI. The initial velocity (Vi) of the phosphorylation reaction was measured at the indicated concentrations of tRNA (left) and ATP (right). Data represent the average values of technical triplicates ± s.d. The Km and Vmax values are shown below each graph.