Fig. 1. Engineering PERSIST-OFF- and ON- motifs for regulated gene expression.
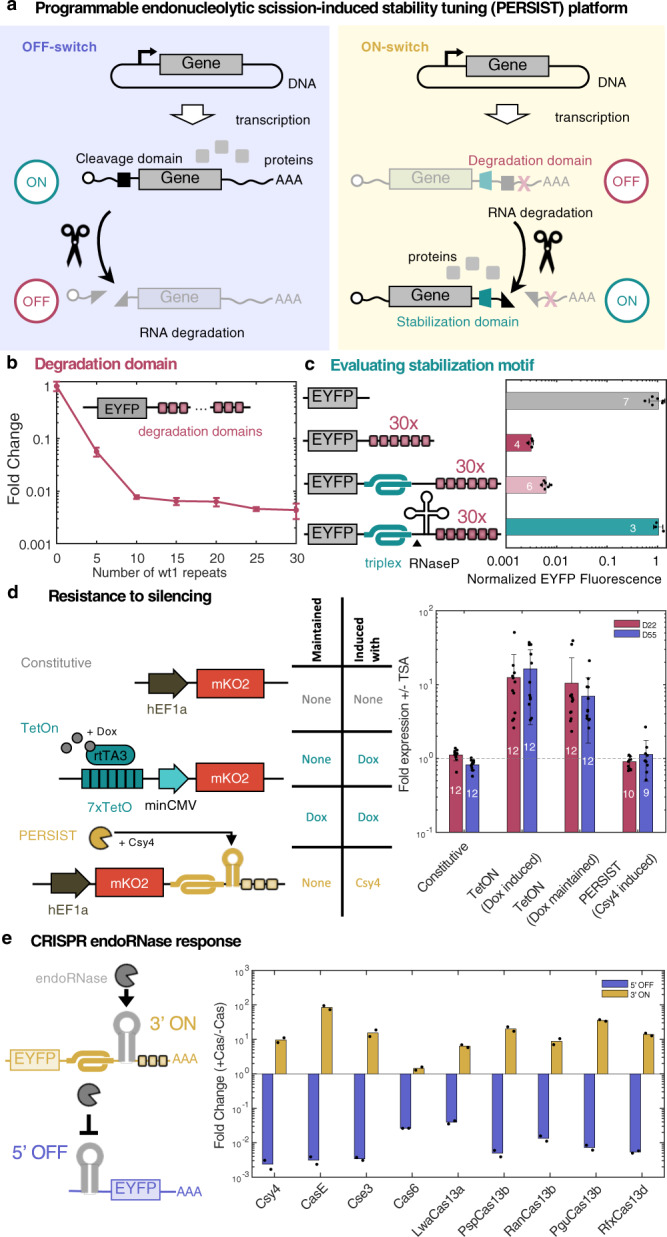
a Schematic of RNA-based OFF-switch and ON-switch motif designs that are regulated by RNA cleavage. b Gene expression reduction due to appending increasing numbers of degradation motifs. Fold change was calculated as described in Supplementary Fig. 3. n = 3 biologically independent samples where each sample represents the evaluation of >1000 transfected cells (HEK293FT). Data are presented as mean ± s.d. c Triplex structure rescues gene expression only after removal of degradation motifs by RNase P. Data were analyzed from the total number of samples indicated in the bars where each sample represents the evaluation of >1000 transfected cells (HEK293FT). Data are presented as mean ± s.d. d PERSIST shows resistance to epigenetic silencing in clonal populations. Three different constructs (left) each expressing mKO2 (constitutive, Tet-On switch, or PERSIST-ON switch) were genomically integrated and single-cell sorted to evaluate clonal silencing effects (see Supplementary Fig. 5). The table (middle) shows the maintenance and induction conditions of each cell line. 22 and 55 days after sorting 12 clonal lines for each cell type were induced with either 4 μM Dox or transfected with Csy4 both in the presence and absence of TSA and evaluated by flow cytometry two days later. The ratio of ±TSA for mKO2 expression was calculated for each cell line in each sample type where a ratio >1 indicates rescue of response by TSA. The Tet-On system shows increased mKO2 expression in the presence of TSA, which indicates epigenetic silencing effects. The PERSIST switch resists silencing similar to the constitutively expressed reporter. Data were analyzed from the total number of clones indicated in the bars where each sample represents the evaluation of ≥6 CHO-K1 cells. Data are presented as mean ± s.d. e CRISPR endoRNases activate the PERSIST-ON motif and repress the PERSIST-OFF motif. Fold change is normalized to the effect of adding endoRNase to a control reporter without an endoRNase recognition site as described in Supplementary Fig. 4. Constructs and endoRNases were optimized (see Supplementary Figs. 6–8; the best variants are shown here). n = 2 biologically independent samples where each sample represents the evaluation of >1000 transfected cells (HEK293FT). Data bars are presented as mean.
