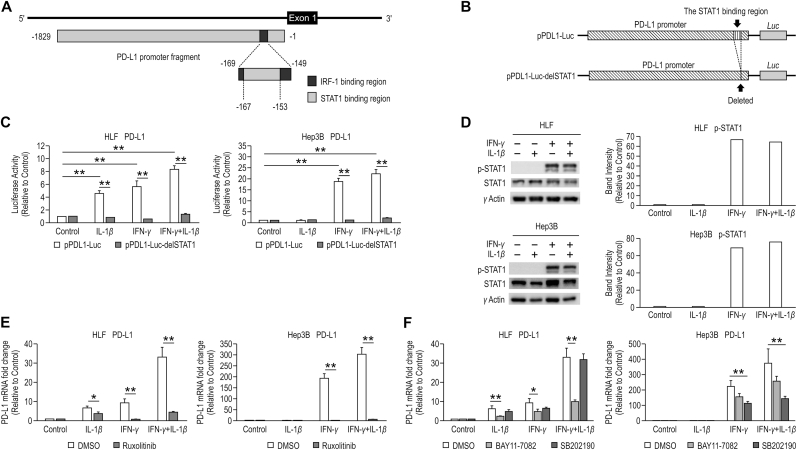
Fig. 2.
STAT1, NF-κB, and p38 MAPK are involved with the synergistic effect of IFN-γ and IL-1β on PD-L1 expression. (A) Diagrammatic representation of the PD-L1 regulatory element. The numbers annotated below the fragments are positions relative to the PD-L1 transcription start site. (B) Diagram of luciferase vectors. The GL4.17 vector with the PD-L1 promoter cloned upstream of the luciferase gene (pPDL1-Luc), and the PD-L1 promoter excluding the STAT1 binding site cloned upstream of the luciferase gene (pPDL1-Luc-delSTAT1). (C) PD-L1 mRNA expression was examined using reporter assays in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 24 h. Firefly luciferase activity was normalized by Renilla luciferase activity. (D) Activation of STAT1 was analyzed by western blotting (WB) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β for 15 min. γ-actin is used as a loading control in WB. The results were expressed as relative ratios of band intensity between pSTAT1 and γ-actin. (E) PD-L1 mRNA expression was examined using quantitative reverse transcription polymerase chain reaction (qRT-PCR) in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β in the presence of dimethyl sulfoxide (DMSO) and Ruxolitinib (1 μM). The samples treated without IFN-γ or IL-1β were used as controls. (F) PD-L1 mRNA expression was examined using qRT-PCR in HLF and Hep3B cells treated with IL-1β (10 ng/mL), IFN-γ (20 ng/mL), or the combination of IFN-γ and IL-1β in the presence of DMSO, BAY11-70825 (5 μM), and SB202190 (5 μM). The samples treated without IFN-γ or IL-1β were used as controls. *P < 0.05, **P < 0.01.