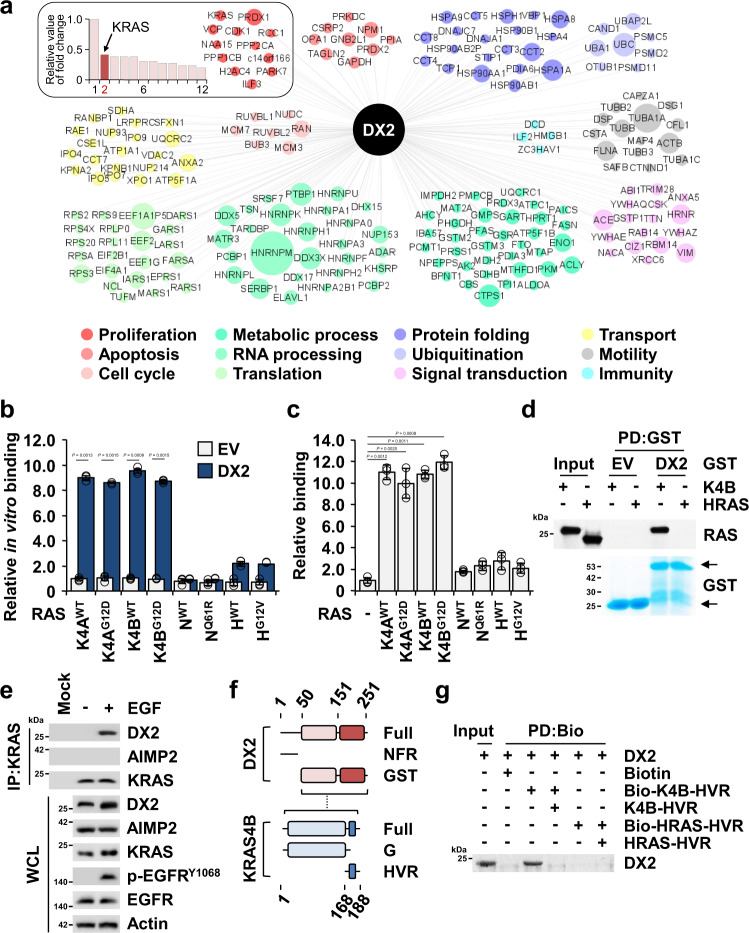
Fig. 2. Binding of DX2 to KRAS.
a Top 200 proteins identified as the potential binding proteins of DX2 by LC-MS analysis. The relative fold change values of 12 proteins in proliferative ontology were represented as the bar graph (left upper box). Among 12 identified proteins, PRDX1 was detected at the highest fold change and the fold changes of other proteins were divided by that of PRDX1. The relative values of the other eleven proteins were graded, taking the value of PRDX1 as 1. Diameter of circles denotes the relative fold changes of a total of 200 identified proteins. b, c Determination of specific binding of DX2 to KRAS. DX2 was mixed with different nanoluciferase-RAS isoforms. Amounts of the indicated RAS isoforms co-precipitated with DX2 were determined by luciferase activity and are shown as a bar graph (b). CCD18CO cells expressing Strep-tagged DX2 and the indicated nanoluciferase-RAS isoforms were precipitated using a strep-tactin column. Amounts of the indicated nanoluciferase-RAS isoforms co-precipitated with DX2 were measured and shown as above (c). All the experiments were independently repeated thrice and error bars denote S.D. Data are presented as mean values ± S.D. P value is from the two-sided t test. d In vitro pull-down assay showing direct binding between DX2 and KRAS4B. Co-precipitated KRAS4B or HRAS with GST-DX2 was detected by SDS-PAGE and immunoblotting using an anti-pan RAS antibody. GST proteins were detected by Coomassie staining. Results are representative of at least three independent experiments. e Endogenous KRAS in DLD-1 cells treated with EGF for 30 min was precipitated with the anti-KRAS antibody. p-EGFR was used as a positive control for EGF signaling. Results are representative of at least three independent experiments. f Schematic diagram presenting the binding region between DX2 and KRAS4B. Binding region is depicted as a dashed line (Supplementary Fig. 4a). g In vitro pull-down assay showing direct interaction between DX2 protein and KRAS4B, but not with HRAS, HVR peptide. Biotin-conjugated and native peptide were serially mixed with DX2 proteins for checking the binding specificity. Results are representative of at least three independent experiments. EV, PD, IP, and WCL represent empty vector, pull-down, immunoprecipitation, and whole cell lysate, respectively. Source data are provided as a Source data file.