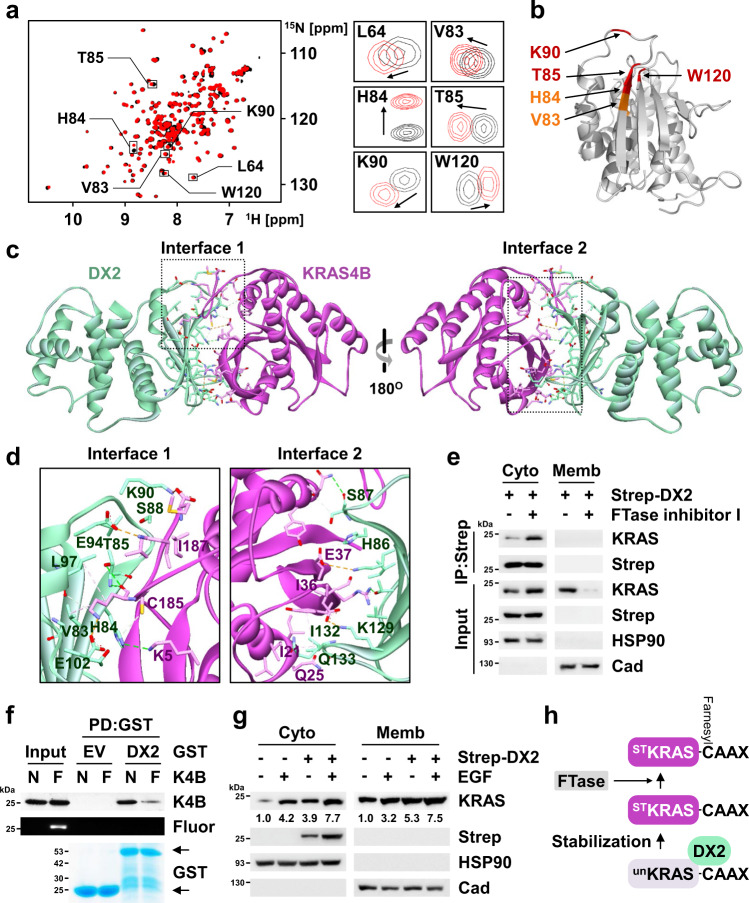
Fig. 3. Analysis of the binding mode of action between KRAS and DX2.
a, b Chemical shift perturbation of 15N-labeled DX251-251(C136S, C222S) by the binding to KRAS4B HVR peptide. Superposition of the 2-dimensional (2D) 1H-15N TROSY spectra of free DX2 in the presence (red) and absence (black) of HVR peptide. Strongly affected residues are depicted (a, left) and the indicated signals are enlarged and presented (a, right). The residues showing strong (Δδ > 0.02 ppm) and medium (0.01 < CSP ≤ 0.02 ppm) perturbation are shown in red and orange, respectively, on the surface of the DX2 GST domain (b). c Result of 600 ns molecular dynamics (MD) simulation to determine the binding surface of DX2 and KRAS4B. The two significant interfaces are highlighted by the dashed box. d Significant residues for binding between DX2 and KRAS4B were analyzed by immunoprecipitation using alanine mutants (Supplementary Fig. 5n, o) and presented in the enlarged image of each interface. e, f Validation of the effect of KRAS4B farnesylation on its binding with DX2. H460 cells expressing Strep-DX2 were treated FTase inhibitor I and separated into cytosol and membrane fraction. HSP90 and cadherin (Cad) were used as loading markers for cytosol (Cyto) and membrane (Memb), respectively (e). In vitro pull-down assay showing the binding of DX2 to farnesylated KRAS4B. In vitro farnestlyated KRAS4B proteins were mixed with the purified GST-DX2. Binding of the two proteins and farnesylation were determined by immunoblotting and fluorescent scans, respectively. N and F indicate native and farnesylation, respectively (f). Results are representative of at least three independent experiments. g H460 cells introducing Strep-DX2 were treated with EGF and separated into Cyto and Memb fractions. Relative levels of KRAS were presented below the KRAS blots. Results are representative of at least three independent experiments. h Schematic presentation for binding between DX2 and KRAS. unKRAS and STKRAS mean unstable and stable KRAS, respectively. Source data are provided as a Source data file.