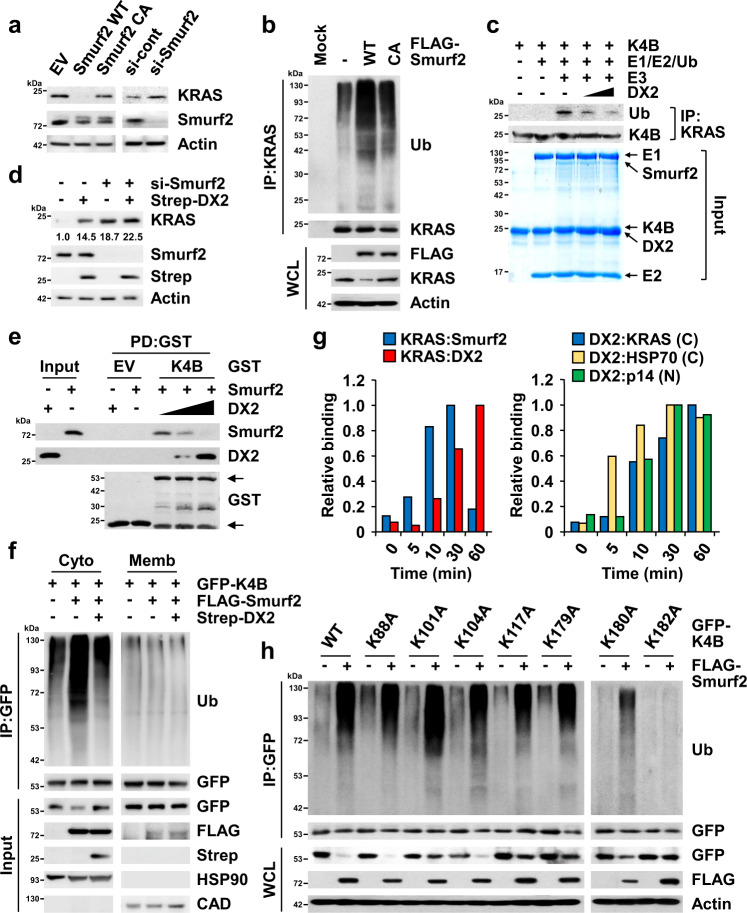
Fig. 4. Competitive blocking of Smurf2-mediated ubiquitination of KRAS by DX2.
a Smurf2-mediated levels of KRAS. Endogenous levels of KRAS from H460 cells expressing Smurf2 wild type (WT), catalytic-inactive mutant (CA), and si-Smurf2 were determined by immunoblotting. si means siRNA. b Smurf2-mediated ubiquitination of KRAS. The cells described in (a) were treated with MG-132 and the ubiquitinated amounts of KRAS were analyzed by ubiquitination assay using the anti-ubiquitin (Ub) antibody. c In vitro ubiquitination assay showing the effect of DX2 on Smurf2-mediated ubiquitination of KRAS4B. UBE1, UbcH5c/UBE2D3, Smurf2, and KRAS4B were used as E1, E2, E3, and substrate for the reaction, respectively. Proteins were confirmed by Coomassie staining in the input panel (bottom). Ubiquitination of KRAS4B was detected by immunoblotting as above (upper). d H460 cells expressing si-Smurf2 and Strep-DX2 were subjected to immunoblotting. Quantitative levels of KRAS were depicted below its blot. e In vitro pull-down assay showing the effect of DX2 on the binding of Smurf2 to KRAS4B. Smurf2, GST-KRAS4B, and DX2 proteins were mixed, and co-precipitation of DX2 and Smurf2 with KRAS4B was monitored. f Inhibitory effect of DX2 on Smurf2-mediated ubiquitination of KRAS4B. 293T cells expressing GFP-KRAS4B, FLAG-Smurf2, and Strep-DX2 were treated with MG-132 and separated into cytosol and membrane fractions. g Analysis of binding kinetics for DX2 or Smurf2 to KRAS (left), and KRAS, HSP70 or p14ARF to DX2 (right) upon EGF signal. All the interactions analyzed by immunoprecipitation (Supplementary Fig. 9a, b) were quantified and presented as graphs. C and N indicate cytosol and nucleus, respectively. The maximum blot intensities obtained from immunoprecipitation of each protein pair was taken as 1, and the other blot intensities were divided by the maximum intensity values and presented as the relative values. h Ubiquitination assay validating the Smurf2-dependent ubiquitination sites of KRAS4B. H460 cells expressing GFP-KRAS4B mutants and FLAG-Smurf2 were treated with MG-132 and subjected to the ubiquitination assay. Source data are provided as a Source data file. a–f, h, Results are representative of at least three independent experiments.