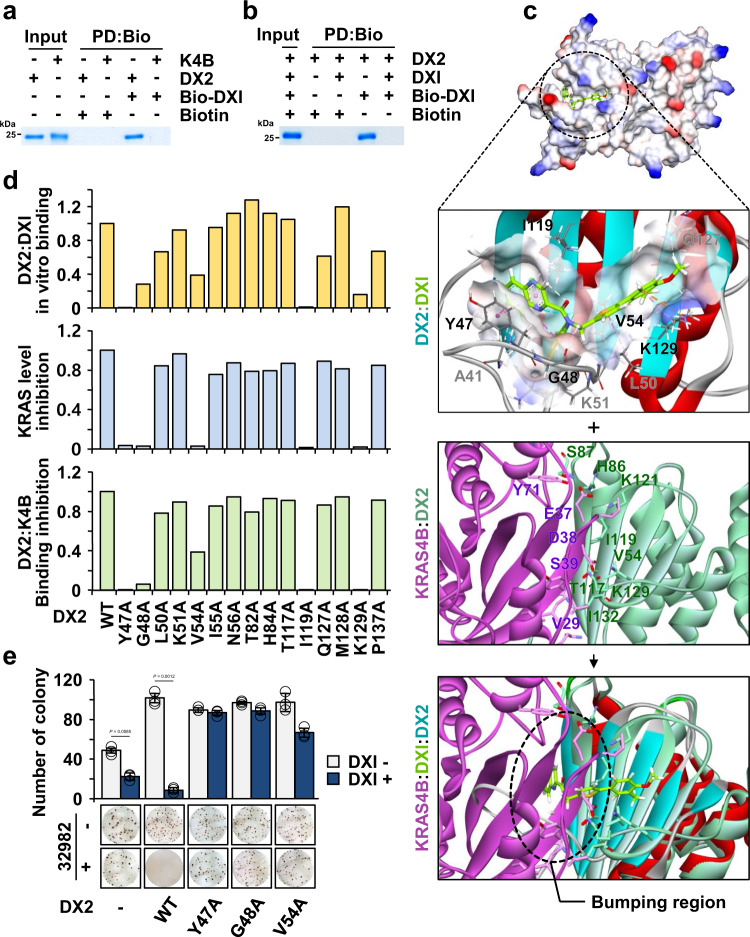
Fig. 6. Mode of action of BC-DXI-32982.
a In vitro pull-down assay showing the direct binding of DXI with DX2, but not with KRAS4B. Proteins co-precipitated with Biotin-DXI #2 (Bio-DXI) were determined by SDS-PAGE and Coomassie staining. Biotin was used as a negative control. Results are representative of at least three independent experiments. b In vitro pull-down assay showing the specific interaction of compound with DX2 using Bio-DXI and DXI. Results are representative of at least three independent experiments. c Binding mode of DXI to DX2 as obtained from the MD simulation. The representative structure at 371.2 ns is shown as an electrostatic surface model (first from above). Zoomed-in view of the detailed interactions is presented and binding residues of the compound (green) were depicted as a stick model (second from above). MD snapshots of DX2 and KRAS4B complex at 533.7 ns (third from above). Overlay of above two MD snapshots by superimposing over DX2. The dashed circle denotes the contact region proposed to be responsible for the DXI-mediated inhibition of the interaction of DX2 with KRAS4B (bottom). d Graphs showing the critical residues of DX2 for binding with DXI. Results from the in vitro binding of DX2 mutants with DXI; compound-mediated change in KRAS levels and inhibition of DX2 mutants-KRAS4B were quantified (Supplementary Fig. 13n–p). e Anchorage-independent colony formation assay validating the significance of the binding between DXI and DX2. Representative images are shown (bottom). The experiment was independently repeated thrice and error bars denote S.D. Data are presented as mean values ± S.D. P value is from the two-sided t test. Source data are provided as a Source data file.