Abstract
MicroRNA (miRNA) is a small, single-stranded, non-coding RNA molecule that plays a variety of key roles in different biological processes through post-transcriptional regulation of gene expression. MiRNA has been proved to be a variety of cellular processes involved in development, differentiation, signal transduction, and is an important regulator of immune and autoimmune diseases. Therefore, it may act as potent modulators of the immune system and play an important role in the development of several autoimmune diseases. Immune thrombocytopenia (ITP) is an autoimmune systemic disease characterized by a low platelet count. Several studies suggest that like other autoimmune disorders, miRNAs are deeply involved in the pathogenesis of ITP, interacting with the function of innate and adaptive immune responses. In this review, we discuss emerging knowledge about the function of miRNAs in ITP and describe miRNAs in terms of their role in the immune system and autoimmune response. These findings suggest that miRNA may be a useful therapeutic target for ITP by regulating the immune system. In the future, we need to have a more comprehensive understanding of miRNAs and how they regulate the immune system of patients with ITP.
Keywords: microRNA, immune thrombocytopenia, gene regulation, autoimmunity, immune, autoimmune diseases
Introduction
ITP is an autoimmune disease characterized by a low platelet count. Most patients have no symptoms or have mild skin bleeding, and 5% of the patients have severe bleeding. 1 Regardless of bleeding, patients with ITP often report fatigue and a decline in quality of life. 2 Overall, the incidence of primary ITP in adults is 2 to 4 per 100000 person-years.3,4 The pathophysiology of ITP is complex and not fully understood. The conventional wisdom is that ITP is mainly caused by autoantibodies against platelet membrane glycoproteins, such as GPIIb/IIIa and GPIb proteins. 5 Antibody-coated platelets are destroyed prematurely in the spleen and/or liver.6,7 T cell-mediated platelet destruction and abnormal number and function of megakaryocytes are also involved.8–11
In recent years, people are more and more interested in exploring the epigenetic mechanism of common diseases, and the number of citations related to microRNA (miRNA) is increasing rapidly. The full picture of the miRNA of these abnormal communications in ITP is slowly being discovered. In this review, we summarize the current knowledge of miRNA in ITP. This work attempt to review some of these studies and summarize some of the latest advances in the role of miRNA in pathophysiology, diagnosis and treatment of ITP.
Biogenesis of microRNA
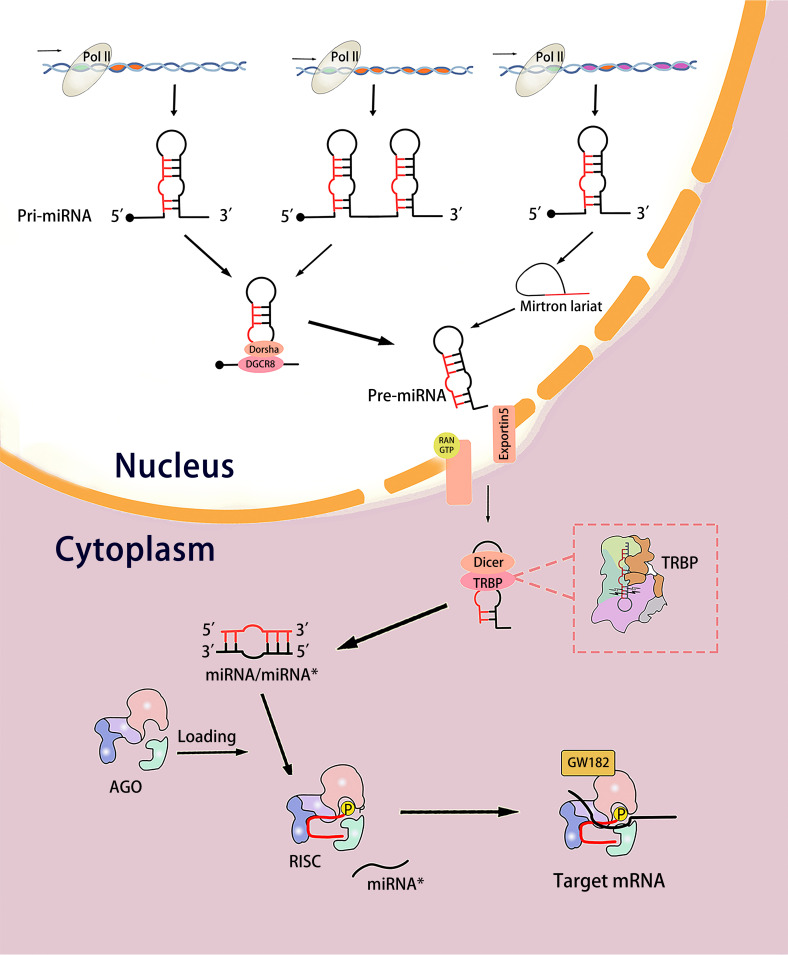
MiRNA is a non-coding RNA of about 22 base pairs (19-25 base pairs in length). MiRNA results in mRNA degradation and translation inhibition by pairing with partial complementary sequences in the 3′ untranslated region (3′UTR) of the target mRNA. Many biological processes of gene expression are regulated by miRNA in this way.12–14 In 1993, two small lin-4 transcripts, approximately 22 and 61nt, were identified respectively. They contain sequences complementary to a repetitive element in the 3′UTR of lin-14mRNA, which proved that lin-4 regulates the translation of lin-14 via an antisense RNA-RNA interaction. 15 MiRNA biogenesis is a complex process that starts from the cell nucleus and consists of two major steps: Transcription of miRNA genes into primary miRNA (pri-miRNA) and pri-miRNA processing through successive cutting steps to maturity (Figure 1).
Figure 1.
The process of miRNA biogenesis. In the nucleus, the miRNA gene is transcribed into pri-miRNAs by RNA polymerase II. Pri-miRNA is cleaved into pre-miRNA through Drosha pathway or Drosha-independent pathway and transported out of the nucleus by Exportin-5 and RAS-related nuclear proteins. In the cytoplasm, pre-miRNA is cut into miRNA/miRNA* by Dizer. Subsequently, the double strands were loaded into the AGO, the miRNA* strand was discarded, while the “guide” miRNA was retained in the AGO to mediate the translation inhibition of the target mRNA.
The miRNA genes are located in the whole genome, most of them come from intergenic regions or antisense to form independent transcriptional units, and most of the others are located in the intron regions of protein-coding genes.16–18 Most intron miRNAs share a promoter with the host gene, some of which have their own promoter and are independently transcribed, while others are transcribed as a single primary miRNA.19,20 Most miRNA genes are transcribed by RNA polymerase II into large pri-miRNAs containing one or more stem-loop structures.21,22 In the nucleus, the pri-miRNA transcript is cleaved into an about 70-nt stem-loop precursor called pre-miRNA.This process is accomplished by a microprocessor complex composed of Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) or generated directly from the intron region of the splice joint in a Drosha-independent pathway.23–25 Subsequently, pre-miRNA is exported from the nucleus by Exportin 5 and RAS-associated nuclear proteins.26–28 In the cytoplasm, pre-miRNA is cleaved into miRNA/miRNA* double-stranded RNA of about 21nt in length near the loop by endonuclide Dicer. A protein called TAR RNA binding protein (TRBP) guides the Dicer cleavage process.29,30 Subsequently, the small RNA double strands produced by Dicer are loaded onto the Argonaute (AGO) protein, and the Hsc70-Hsp90 chaperone promotes the conformational change of AGO, thereby allowing the loading of miRNA double strands to form RNA-induced silencing complex (RISC).31–33 The AGO complex that binds to the RNA double-strand is called pre-RISC. Usually, the two chains have different fates. The “passenger” chain is discarded, while the “guide” (mature) miRNA is retained in the AGO protein. There are several possible mechanisms to remove passenger strand. If the double strands match in the center, the AGO2 protein can cleave the passenger chain and be further degraded by the endonuclease C3PO to promote RISC activation.34,35 But most miRNAs have a central mismatch, and the AGO family (AGO1-4) has no cutting ability except AGO2, so double strand dissociation is a more common way.36–40 The mismatch of nucleotide positions 2-8 and 12-15 of the guide chain promotes the dissociation of miRNA double strands.41–44 This asymmetric chain selection (often referred to as the “asymmetry rule”) depends on the relative thermodynamic stability of the first 1-4 bases at both ends of the small RNA double strand. 45 The unstable chain at the 5′ end is selected as the guide chain, while the other chain is discarded.46–48 At this point, a functional RISC, which is called a mature RISC is formed. This ribo-nuclear protein complex mediates the translational repression of target mRNA followed by deadenylation and decay.49–51 MiRNA is the nucleic acid core of mature RISC, and its sequence determines which mRNAs in a given transcriptome it may interact with, but it is the protein components of mature RISC that perform target mRNA silencing. If the target is completely complementary in the base pairing region, it is cleaved by the catalytic active AGO. However, when the target is not completely complementary, AGO is not enough to silence, so it is necessary to recruit from the GW182 family proteins. GW182 protein acts as a molecular scaffold to connect AGO protein and downstream effect complex in mammalian cells, which mediates miRNA-dependent translation repression and deadenylation.12–14
New discoveries have been made in the research on the biogenesis and function of miRNA. Emerging evidence also suggests that miRNA is associated with the pathogenesis of human diseases such as cancer and metabolic disorders.
miRNA and Autoimmune Diseases
Up to now, many clinical studies and experimental animal models have proved that miRNA is involved in the regulation of immune system homeostasis and is involved in the pathogenesis, diagnosis and prognosis of many autoimmune diseases.52–54 The inability of the autoimmune system to distinguish between the self and the non-self is often referred to as violation of tolerance, which is the basis of autoimmune diseases. 55 The disease cluster includes more than 100 diseases, ranging from a single organ, such as ITP, type 1 diabetes, and autoimmune thyroiditis, to systemic involvement, such as systemic Lupus Erythematosus (SLE), rheumatoid arthritis (RA). The study of abnormal expression of miRNA in a variety of autoimmune diseases has been widely carried out, and showed a correlation. For example, miR-21 is up-regulated in patients with SLE, asthma and RA;56,57 miIR-326 is overexpressed in Multiple sclerosis, type 1 diabetic patients with ongoing islet autoimmunity, SLE, but decreased in ITP;58,59 MiR-155 is up-regulated in ITP but down-regulated in Guillain-Barre syndrome. 60 Interestingly, the expression of miRNA is different in different cell or tissue types derived from RA, for example, the expression of miR-155 is increased in synovial tissue, RA synovial fluid, peripheral blood B cells, peripheral blood macrophages and synovial fluid macrophages, but decreased in plasma and serum. 61 Existing studies have shown that miRNA has similarities and contradictions in autoimmune diseases, because the mechanism of miRNA is not completely clear. One miRNA can target hundreds of mRNAs, and the expression of an mRNA is regulated by multiple different miRNAs. This bidirectional multiline regulatory network may play an important role in autoimmune diseases.
MiRNAs Abnormally Expressed in ITP
In ITP, the association between miRNA and immune interaction is one of the most interesting research areas that has been neglected in recent years. MiRNA affects almost all immune cells and immune events. How does miRNA change in patients with ITP? More and more studies have given the answer. In this review, we try to give only a brief overview of the latest findings (The main microRNAs shown in Table 1).
Table 1.
The Main MicroRNAs# Involved in the Different Cells of ITP.
| MicroRNA | Species/Sample source | Target Gene | Function | MiRNA Regulation | References |
|---|---|---|---|---|---|
| miR-130a | Human/PBMCs | TGFB1 IL-18 |
- | Downregulated | 62 |
| miR-409-3p | Human/PBMCs | IFNG | - | Downregulated | 63 |
| miR-106b-5p | Human/PBMCs | - | - | Upregulated | 64 |
| Human/CD4+ T | NR4A3 | Regulate immune imbalance of Treg/Th17 through the NR4A3/Foxp3 pathway | Upregulated | 64,65 | |
| miR-200C-3p | Human/PBMCs | - | - | Upregulated | 64 |
| miR-92a-3p | Human/PBMCs | - | - | Upregulated | 64 |
| miR-142-3p | Human/PBMCs | - | - | Upregulated | 64 |
| Human/Treg | - | - | Downregulated | 66 | |
| Human/PBMCs | - | - | Downregulated | 67 | |
| miR-146b-5p | Human/Treg | - | - | Downregulated | 66 |
| miR-146a | Human/PBMCs | - | - | Downregulated | 67 |
| miR-146a-5p | Human/CD4+ T cells | - | Contribute to the Th17/Treg imbalance | had no significant difference | 68 |
| Human/BMSCs-exosome form health volunteer | IRAK | Regulate Th17/Treg imbalance | Upregulated | 69 | |
| miR-326 | Human/PBMCs | - | - | Downregulated | 67 |
| Human/CD4+ T cells | - | Contribute to the Th17/Treg imbalance | Had no significant difference | 68 | |
| miR-17-5p | Human/PBMCs | - | - | Upregulated,but had no significant difference | 67 |
| Human/CD4+ T cells | - | Contribute to the Th17/Treg imbalance | had no significant difference | 68 | |
| miR-181a | Human/PBMCs | - | - | Upregulated,but had no significant difference | 67 |
| Mouse/Macrophage | - | Regulate of TLR4, TNF-α and IL-6 expression | Downregulated (by a TCM treatment) | 70 | |
| miR-181-5p | Human/CD4+ T cells | - | Contribute to the Th17/Treg imbalance | Had no significant difference | 68 |
| miR-21 | Human/PBMCs | - | - | Upregulated,but had no significant difference | 67 |
| miR-155 | Human/Treg | - | - | Downregulated | 66 |
| Human/PBMCs and macrophages | SOCS1 | Role in macrophage M2 polarization | Upregulated | 71 | |
| Human/PBMCs | - | - | Upregulated | 72 | |
| Human/PBMCs | SOCS1 | Regulate cytokine profiles | Upregulated | 73 | |
| Human/CD19+ B cells | - | Role in the dysfunction of B lymphocytes and ntracellular antibody production | Upregulated | 74 | |
| Human/CD4+ T cells | - | Contribute to the Th17/Treg imbalance | Has no significant difference | 68 | |
| miR-15a | Human/PBMCs | - | Involve in the regulation of Th1/Th2 imbalance | Downregulated | 75 |
| Let-7b-5p | Human/B cells | BAFF-R | Enhance the expression of surface BAFF-R and promotes B cell survival | Upregulated | 76 |
| miR-99a | Human/CD4+ T cells | mTOR | Contribute to the Th17/Treg imbalance | Downregulated | 68 |
| miR-182-5p | Human/CD4+ T cells | - | Contribute to the Th17/Treg imbalance | Upregulated | 68 |
| miR-183-5p | |||||
| miR-96-5p | |||||
| miR-125-5p | Human/CD4+ T cells | CXCL13 | Induce immune imbalance of Treg/Th17 | Downregulated | 77 |
| miRNA-30a | Mouse/Spleen mononuclear cells | SOCS3 | Affect the differentiation of Th17 cells | Upregulated | 78 |
| miR-199a-5p | Mouse/spleen CD4+ T cells | STAT3 | Represse Th17 differentiation | Downregulated | 79 |
| miR-98-5p | Human/BMSCs | IGF2BP1 | Exert Pro-apoptotic Effects by Inhibiting the IGF-2/ PI3K/Akt Pathway | Upregulated | 80 |
| miR-20b-5p | Human/BMSCs | - | - | Upregulated | 80 |
| miR-3148 | |||||
| miR-4284 | |||||
| miR-3977 | |||||
| EBV-miR-BART19-3p | Human/Megakaryocytes | - | - | Upregulated | 81 |
| HSV1-miR-H7* | |||||
| HSV1-miRH8* | |||||
| HSV1-miR-H14-3 | |||||
| HSV2-miR-H7-3p | |||||
| MiR-557 | Human/Plasma | - | Role in the excessive apoptosis of megakaryocytes | Upregulated | 82 |
| miR-338-5p | Human/Platelet | - | - | Only expressed in ITP | 83 |
| miR-765 | |||||
| miR-122-5p | |||||
| miR-451b | |||||
| miR-133a-3p | Downregulated | ||||
| miR-224-3p | |||||
| miR-452-5p | |||||
| miR-491-5p | |||||
| miR-15a-3p |
Note:# MicroRNAs measured by quantitative PCR.
ITP: immune thrombocytopenia; PBMCs: peripheral blood mononuclear cells; BMSCs: bone mesenchymal stem cells; TCM: Traditional Chinese medicine.
Mononuclear Cell
Human peripheral blood mononuclear cells (PBMC) are an important part of the immune system and a natural defense against infection and foreign invaders. PBMC is a mixture of lymphocytes (T cells, B cells and NK cells), monocytes and dendritic cells, of which lymphoid cells are the most abundant, accounting for 70-90%. 84 Like the plasma separation method, the PBMC separation method is simple and standardized, and the quality of the samples obtained is stable. PBMC is widely used in ITP research.
Several studies have examined the miRNAs expression profile of PBMC in patients with ITP. In 2007, Dai and his colleagues identified miRNA in PBSCs from ITP and SLE patients using microarray analysis. 85 It was found that 19 miRNAs related to ITP, 14 miRNAs of them were down-regulated and 5 miRNAs were up-regulated, but most of them were not verified by quantitative or semi-quantitative experiments. The expression of HSA-miR-299-3p and MMU-miR-298 is up-regulated in SLE and down-regulated in ITP, which provides a possible biomarker for the differentiation of ITP and SLE. This is the earliest literature we have retrieved about miRNA in patients with ITP. Due to the rapid development of technology, the results of this study need to be reconsidered.
Compared with normal controls the expression of miR-130a and miR-409-3p in PBMC of active ITP patients decreased significantly and recovered after effective treatment. 62
Using the same method, the two studies proved that the transforming growth factor-β 1 (TGFB1) and interleukin 18 (IL-18) genes are the target genes of miR-130a, and IFNG is the target gene of miR-409-3p. 62 Although they are found to be related at the genetic level, two contradictory results that are difficult to explain. First, the expression levels of miR-130a and TGFB1 in plasma decreased; second, the concentration of IFNG in plasma did not change with miR-409-3p before and after treatment. For the decreased expression of TGFB1 and miR-130a, the authors speculate that there is a regulatory feedback loop between TGFB1 and miR-130a, and the decrease of TGFB1 leads to the down-regulation of miR-130a, which partly saves the expression of TGFB1. However, exogenous TGFB1 does not promote the miR-130a expression of PBMCs in any way, which does not support this conjecture. Since platelets are the largest source of TGFB1, thrombocytopenia in patients with ITP may be the main cause of TGFB1 decrease.
IFNG is elevated in the plasma of patients with ITP. 86 MiR-409-3p negatively regulates the expression of IFNG, so the decrease of miR-409-3p in ITP patients may partly explain the increase of IFNG, but it is not the main mechanism. Only when future studies directly verify that both miRNA and cytokines come from PBMCs, these contradictions can be resolved. It is well known that a single miRNA has the ability to regulate multiple genes, so the functions of MIR409-3p and miR-130a may be much more complex than expected. The relationship between the abnormal expression of them and the pathology of ITP needs to be further discussed.
The level of DGCR8 transcription in PBMC of ITP patients was significantly lower than that of healthy controls, and the expression of miR-409-3p was positively correlated with DGCR8 transcripts (r = 0.570,P = 0.005). There was no correlation between other enzymes (DROSHA、DICER1、DGCR8 and EIF2C2) related to miRNA biogenesis. It shows that the reduction of DGCR8 leads to impaired processing of miRNAs from Pri-miRNAs to Pre-miRNAs and ultimately leads to the reduction of miR-409-3p. 63
The expression profiles of miRNAs and mRNAs in PBMC of 3 subjects in each group were detected by gene chip technique. 64 Compared with healthy subjects, there were differences in the expression of miRNAs and mRNAs in newly diagnosed and chronic ITP patients. The miRNA-mRNA regulatory network was constructed through bioinformatics, and it was found that three up-regulated miRNAs (miR-106b-5p, miR-200C-3p, and miR-92a-3p) exhibited strong regulatory abilities, which were regulated more transcription factors in both stages. Verified by qPCR, there were significant differences in miR-106b-5p and miR-200C-3p among the three groups, and there was no significant difference in the expression of miR-92a-3p between the chronic group and newly diagnosed group. The area under the ROC curve of the differential expression of miR-106b-5p between the newly diagnosed group and the healthy control group is the largest (0.92). The difference in expression of miR-106b-5p and miR-200C-3p can distinguish the chronic group and healthy controls (AUC is 0.88). These three miRNAs can effectively distinguish ITP stages. In this study, it was found that the expression of miR-142-3p was abnormal and elevated. Similarly, miR-106b-5p in peripheral blood of patients with ITP is significantly up-regulated, which mediates the immune imbalance of Treg/Th17 in ITP. 65
There is no consistent answer to the expression of miR-155 in ITP. Compared with healthy controls, miR-155 was significantly decreased in Treg cells of ITP patients; Although the expression increased in PBMC and CD4+ T-cells, there was no statistical significance. 66 However, three other studies have shown that the expression of miR-155 is up-regulated in PBMCs of patients with ITP. 71 The relative expression level of miR-155 in PBMCs of elderly patients with ITP increased significantly with the severity of the disease (P < 0.05). The response rate to glucocorticoid of elderly ITP patients with high expression of miR-155 was significantly lower than that of patients with low expression of miR-155 (72.31% vs 96.92%). And patients with high expression have a higher risk of recurrence. 72 Another study suggests that miR-155 may be involved in the pathogenesis of ITP by targeting SOCS1 to regulate cytokine profiles. 73 The miR-155 level was negatively correlated with platelet count, SOCS1 mRNA level and plasma IL-4, IL-10 and TGF- β 1 levels, but positively correlated with plasma IL-17A level. Medical treatment reduced the level of miR-155, but there was no significant difference before and after treatment (p = 0.056). SOCS1 is a mature member of the SOCS family. Some studies have determined that SOCS1 is one of the targets of miR-155 and participates in the function of T cells, macrophages, and dendritic cells.87–90
The expression of miR-15a is significantly reduced in PBMC of patients with ITP. The expression of miR-15a is negatively correlated with IFN-γ and IL-2, and positively correlated with IL-4 and IL-10. Overexpression of miR-15a can significantly inhibit the production of IFN- γ and IL-2, and promote the production of IL-4 and IL-10. Since IFN-γ and IL-2 are inflammatory factors secreted by Th1 cells, and IL-4 and IL-10 are secreted by Th2 cells, it is speculated that miR-15a may be involved in the Th1/Th2 imbalance in ITP. 75
The results of a study using deep sequencing showed that there were 6 miRNAs differentially expressed between ITP and control (P < 0.0005), of which 2 miRNAs (miR-4482-3p and miR-1299) were up-regulated, and the other 4 miRNAs (miR-412-5p miR-204-5p miR-199a-5p and miR-126-5p) were down-regulated. 91
miRNAs and the Innate Immune System
Macrophages
The phenotype and function of macrophages are regulated by the surrounding cytokine environment. Induced by Th1 cytokines like IFN-gamma and microbicidal stimuli such as lipopolysaccharides (LPS), macrophages polarized to M1, produced high levels of pro-inflammatory cytokines(such as IL-12、IL-23), and polarized to M2 induced by Th2 cytokines such as IL-4\IL-13.92,93 M2 high secretion of interleukin-10 (IL-10) has anti-inflammatory properties.94,95 Current studies have shown that splenic macrophages promote platelet destruction in ITP. 96 And macrophages act as antigen-presenting cells to stimulate the acquired immune response. It is generally believed that platelet destruction after autoantibody binding occurs in the spleen and binds to FcgRIIa and FcgRIIa on the reticuloendothelial system tissue macrophages through the Fc portion of the platelet surface immunoglobulin. 97 MiRNA affects all these events, from the activation of macrophages to inflammation and immunity. 98
The expression of miR-155-5p is up-regulated in patients with ITP and affects the inflammatory activity of macrophages. MiR-155-5p can inhibit SOCS1-dependent PD1/PDL1 pathway and promote M2 polarization of macrophages to prevent ITP. Chang et al 71 confirmed that the expression of SOCS1 in macrophages and PBMC of newly diagnosed and treated ITP patients was significantly lower than that of healthy controls, while miR-155-5p was up-regulated. Further exploring the relationship between the two through in vitro experiments and animal models, it was found that miR-155-5p targeted SOCS1 and down-regulated its expression. More specifically, inhibition of miR-155-5p can produce a series of changes: first, promote the expression of CD206 (M2 phenotypic protein) and up-regulate M2/M1; second, up-regulate the expression of PD1/PDL1; third, up-regulate the expression of SOCS1. When SOCS1 is overexpressed in macrophages, the above processes also occurred while the expression of miR-155-5p did not change. The cells that inhibited miR-155-5p and silenced SOCS1 reversed the above process, indicating that SOCS1 is the target of miR-155-5p. In PBMC, the expression of cytokine IL-4\IL-10\TGF-BETA increased and IL-17A decreased after miR-155-5p was inhibited. Another study of PBMCs in patients with ITP showed similar results: miR-155 increased, SOCS1mRNA level decreased in PBMCs of ITP patients, and miR-155 level decreased after medical treatment. 73
The miR-155 host gene (MIR155HG) produces two different miRNA chains, miR-155-5p from 5p precursor and miR-155-3p from 3p precursor. In humans, the highly expressed miRNA tends to 5p hairpin arm, so miR-155-5p is generally considered to be a functional miR-155 form.99,100 MiR-155 is considered to be an important regulator of many acquired and innate immune cell populations.87,89 Studies have shown that compared with M2, MiR-155 is highly expressed in M1 macrophages.101,102 Inhibition of miR-155 can polarize macrophages to M2. 103 There are M1 polarization dominance and M2 marker expression impairment in spleen and peripheral macrophages of patients with ITP, and the proportion of M2 increases after treatment, suggesting that they may be involved in the pathogenesis of ITP.104,105 These results suggest that miR-155 is involved in the pathogenesis of ITP by regulating the polarization of macrophage M1/M2.
Another study found that miR-181a was involved in the treatment of ITP. 70 A passively immunized ITP mouse model was established, and the therapeutic effect of Qian Five Rhinoceros Gindeng (QFRG) on ITP mice was observed. In this group, the platelet count and the expression of miR-181a in PBSC increased significantly, while TLR4, IL-6, and TNF-ɑ decreased significantly. Inhibition of miR-181a expression in the model reversed the therapeutic effect. TLR-4/IL-6 and TNF-ɑ decreased significantly after upregulating the expression of miR-181a in LPS-induced mononuclear macrophages with miR-181a mimics in vitro. Bi et al 106 found that the expression of miR-181a increased when the M1 phenotype was transformed into M2. However, more direct evidence is needed to confirm the polarized involvement of macrophages in ITP.
Dendritic cells and Nk cells
As an important part of innate immunity, dendritic cell dysfunction is involved in the disease process of ITP, like the expression of CD80 and CD86 increased, while the expression of CD70 and CD205 decreased significantly.107,108 There are few studies on NK cells in ITP. In patients with ITP, the number of NK cells in peripheral blood decreased, developmental disorders and function decreased, and insufficient inhibition of self-reactive T and B cells led to the occurrence of diseases.109,110 Up to now, there is no direct evidence as to whether microRNA is involved in the reaction of DC cells and NK cells in ITP.
Adaptive Immune System
B-cell
B-cells play an important role in ITP. Plasma from patients with ITP was injected into healthy volunteers, causing severe thrombocytopenia. 111 It was subsequently proved that B cells produced antiplatelet antibodies, mainly IgG antibodies, and a small number of IgM and IgA antibodies could also be detected.112,113 The antibody binds to rich glycoproteins or glycoprotein complexes on the surface of platelets (especially GPIIb/IIIa and GPIb/IX/V, rare GPIa/IIa, IV, or VI).113,114 Phagocytosis of antibody-labeled platelets by splenic macrophages mediated by Fc γ receptor.115,116 The proportion of total B cells, naïve B cells, and defective Bregs decreased in patients with ITP. 117 The loss of peripheral tolerance of immature B cells may be an important part of the pathogenesis of ITP. 118 Based on these immunopathological theories, B cell depletion is an effective strategy for the treatment of ITP.
There are two reports about the abnormality of miRNA in B cells of ITP. Let-7b-5p and miR-155 are abnormally highly expressed in B cells derived from peripheral blood of patients with ITP. 74
Wang et al 76 found that the expression of let-7b-5p in B cells of patients with ITP was up-regulated, and it was higher in patients with positive platelet specific autoantibodies than in patients with negative platelet specific autoantibodies. Compared with healthy controls, the level of B-cell-activating factor receptor (BAFF-R) on the surface of B cells in patients with ITP was higher but the level of BAFF-R mRNA was lower. This inconsistency points to the post-transcriptional regulation of BAFF-R and may be related to miRNA. In order to further investigate whether let-7b-5p regulates BAFF-R expression, the researchers stimulated CD19+ B-cells from healthy volunteers with BAFF, which resulted in an up-regulation of let-7b-5p expression After up-regulation of let-7b-5p in B cells in vitro, the expression of BAFF-R increased and promoted the survival of B cells, while the mRNA level of BAFF-R decreased, which was consistent with the expression of B cells in patients with ITP. These findings suggest that let-7b-5p may enhance BAFF/BAFF-R-mediated signal transduction, thus promoting B cell survival and participating in the disease process of ITP.
B-cell-activating factor (BAFF) is a key cytokine supporting B-cell homeostasis and tolerance. BAFF/BAFF-R interaction determines the fate of mature B cells and plays an important role in the occurrence and development of autoimmunity.119–123 BAFF is significantly increased in circulation in patients with ITP. It is speculated that BAFF may play a pathogenic role in ITP by promoting B cell survival and increasing B-cell effector function, thereby increasing platelet apoptosis and IFN-γ secretion.124,125 BAFF binds to three receptors: TNF, TACI, BCMA, and BAFF-R. BAFF-R is one of the main survival-promoting receptors of BAFF and is widely expressed in human B-cell subsets, especially mature B-cells.126,127 BAFF-dependent survival signals are mediated by NF- κ B activation.104,105 Of the three BAFF receptors, only BAFF receptor (BAFF-R) increased in ITP mice. 128 BAFF-R not only prolonged the survival of B-cells but also significantly promoted the survival of CD8+ T-cells and the proliferation of B-cells in patients with ITP. 129
The let-7 family is the second miRNA found in Caenorhabditis elegans, which is involved in the immune response of several cancers and the regulation of tumor resistance. The expression of let-7b-5p is increased in B-cells of patients with ITP, especially in patients with positive platelet autoantibodies, and let-7b-5p can enhance the expression of BAFF-R. These data suggest that let-7b-5p may be a functional miRNA regulating antibody production and B cell survival.
Another study on CD19+ B-cells found that miR-155 in B-cells of newly diagnosed ITP patients was higher than that of the control group, IgG and IgM, which reflect the activity of B lymphocytes, had similar results, while the results of SHIP1 were opposite to those of miR-155. 74 MiR-155 was negatively correlated with platelet count and positively correlated with the number of peripheral B1 (CD19+ CD5+ ) cells. It is speculated that miR-155 may stimulate the activation of B-cells receptors by silencing the expression of negative regulatory factor SHIP-1 in B-cells, and then over-activate autoreactive B-cells, promote the production of specific antibodies IgG and IgM, and mediate platelet destruction.
SHIP-1, a member of the inositol phosphatase family, plays an important role in the maturation of B lymphocytes and the production of high affinity antibodies. 130 It is known that miR-155 plays a role in hematopoiesis, inflammation and immune response, and is highly expressed in a variety of solid tumors. 131 In B cell proliferative diseases such as diffuse large B-cell lymphoma, the level of miR-155 increases, and the expression of SHIP-1 decreases. Inhibition of SHIP-1 is an important part of the biological function of miR-155.89,132,133 This study proved that, like other diseases, B lymphocytes in patients with ITP had high expression of miR-155 and inhibition of SHIP-1, suggesting that miR-155 can be used as a therapeutic grouping and prognostic molecule for ITP.
T-cell
Abnormal T-cell immunity plays an important role in the pathogenesis of ITP. Excessive activation and proliferation of platelet autoantigen reactive cytotoxic T cells, imbalance of T-cell subsets, and functional abnormalities inhibit megakaryocyte apoptosis. 134 In recent years, it has been found that T cell immune imbalance plays an important role in the pathogenesis of ITP. Many studies have focused on the imbalance of T cell subsets. At present, it is widely accepted that the phenotypes of Th1, Th17 and newly discovered Th9, Th22 and T follicular helper cells increase, while the number of cells with immunomodulatory characteristics (such as T regulatory cells) decreases. 135
Treg cells account for 5-7% of CD4+ T-cells, and they develop directly in the thymus and periphery. 136 Many inhibitory activities of Treg cells can act in an antigenic non-specific way, inhibiting different specific effector T cells. 137 It can also regulate the tissue microenvironment and promote the emergence of other immunosuppressive cell populations by producing some of the same immunosuppressive molecules (such as IL-10, TGF-β, and IL-35). 136 Th17 cells produce the signature cytokine interleukin 17A (IL-17A) and lineagespecific transcriptional factor retinoid-related orphan receptor gamma t (RoR-γt).138,139 One of the most striking aspects of Treg physiology is their opposition to pro-inflammatory Th17 cells. 140 Th17 cells cause autoimmunity, while Treg cells inhibit autoimmunity. Many factors affecting the production and maintenance of Treg and Th17 cells are also important for the regulation of their balance, including TCR signal, costimulatory signal, cytokine signal, Foxp3 stability and, metabolic process. 141 Th17 and Treg developmental programs are interrelated, and immature T-cells develop to Th17 or Treg phenotypes in different extracellular environments.142,143 The altered homeostasis between Th17 and Treg has been implicated associating with several autoimmune diseases.144–147 In patients with ITP, the number of Tregs decreases or shows a deficiency of inhibitory function, while the number of Th17 cells increased and hyperfunction. 148
MiRNA regulates Treg/Th17 balance in ITP, and more than 10 miRNAs are found to be involved in experimental animal models and clinical specimens (miR-30a miR-199a-5p miR-106b-5p miR-155-5p miR-156b-5p miR-142-3p miR-99a miR-182-5p miR-183-5p miR-125a-5p miR-146a miR-326 miR-142-3p).
Genome-wide expression analysis of mRNA and microRNA in T-cells of ITP patients and controls showed that there were 1915 genes and 22 miRNAs differentially expressed. 149 Then TargetScan and Miranda databases were used to predict target genes, of which 17 microRNA were related to changes in target gene expression. According to the functional classification of cross-reference genes by GO, 57 of these target genes are related to the immune system, including T cell activation involved in immune response, natural killer cell differentiation, regulation of immunoglobulin production, positive regulation of leukocyte activation, lymphocyte activation involved in immune response, lymphocyte differentiation and lymphocyte costimulation. The researchers found that plasma levels of CXCL13 and IL-21 in patients with ITP were significantly higher than those in the control group. 149 This study opened the beginning of the study of T-cell miRNA in patients with ITP. In the subsequent scientific research results, there are more and more studies on miRNA in T-cells of patients with ITP.
Hua et al 68 found that miR-99a regulates the differentiation of Treg cells by targeting mTOR in ITP.Compared with the healthy control group, the expression of miR-99a in CD4+ T cells of ITP patients was down-regulated. The percentage of Treg cells, and the decrease of plasma IL-10 in ITP patients were positively correlated with the expression of miR-99a. There was no significant correlation between the expression of miR-99a and TGF-β1. As mentioned earlier, IL-10 and TGF-β are cytokines secreted by Treg. Up-regulation of miR-99a with mimics promotes CD4+ cells to differentiate into Treg, and the expression of mTOR and its phosphorylation level is decreased. On the other hand, the study also found increased expression of miR-182-5p and miR-183-5p on CD4 cells and increased TH17 cell ratio in ITP patients. Down-regulation of miR-183-5p inhibited Th17, while down-regulation of miR182-5p had no effect on Th17 differentiation. IL-17, a Th17-related cytokine, was overexpressed in ITP patients, but not significantly associated with miR-182-5p or miR-183-5p. MiR-183-5p may be used as a molecular basis for the classification of ITP because the expression of miR-183-5p in CD4+ T-cells of critically ill patients is significantly higher than that of non-critically ill patients. It was found that there was no significant difference in the expression of miR-146a-5p,miR-155-5p,miR-96-5p,miR-17-5p,miR-181-5p, and miR-326 in CD4+ T cells between patients and healthy controls, and there was no significant effect on the regulation of Th17 and Treg cell differentiation.
Different from the results of Hua et al, miR-146a-5p,miR-155-5p, and miR-326 obtained different results in the other two groups of studies.
Liu et al 67 studied the expression profiles of 7 kinds of immune-related miRNAs (miR-155, miR-146a, miR-326, miR-142-3p, miR-17-5p, miR-21 and miR-181a) and four kinds of Th cells (Th1, Th2, Th17 and Treg) in PBMC of ITP patients. The results showed that the expression of miR-146a,miR-326, and miR-142-3p in PBMC of ITP patients was significantly lower than that of healthy controls. The expression of MiR-142-3p and miR-146a was negatively correlated with Th17 cells, while the expression of miR-146a was positively correlated with Treg cell frequency and platelet count, respectively. Dexamethasone-treated PBMCs can up-regulate the expression of miR-146a. Th17 frequency decreased after miR-146a was up-regulated with agomir. MiR-155, miR-17-5p, miR-21, and miR-181a were not found to be statistically significant. There was no significant difference in miR-17-5p and miR-155 between ITP patients and healthy controls, which was consistent with the results of Hua et al, however, the results of miR-146a, miR-326, and miR-181a expression were not consistent, which may be due to the different cell phenotypes of the studies.
Zhu et al 66 found that miR-155-5p, miR-146b-5p and miR-142-3p decreased in Treg cells of ITP patients. The researchers first used miRCURY LNA array to evaluate the expression of human miRNA in T cells of 3 ITP patients and 3 healthy blood donors, then further analyzed 37 genes (fold > 1.3 and p < 0.05) and found that 26 genes were up-regulated and 11 down-regulated. After qPCR verification, miR-155p, miR-146b-5p and miR-142-3p in Treg were significantly down-regulated in ITP patients.
In the third case, the expression of miR-155 in ITP CD4+ cells and PBMCs was increased but not significantly different from that in healthy controls, while it was significantly decreased in the Treg group.67,68 As for miR-142-3p and miR-146, they were down-regulated in PBMCs and Treg. But what is less certain is whether the functions of miR-146a and miR-146b are consistent in the pathophysiology of ITP. Although there are only two bases different between the two.150,151
Recent studies have found that miR-106b-5p negatively regulates NR4A3/Foxp3 and mediates the immune imbalance of Treg/Th17 in ITP. 65 Compared with healthy controls, miR-106b-5p was significantly up-regulated and NR4A3 mRNA was significantly down-regulated in peripheral blood of patients with ITP, and there was a negative correlation between them. TGF-β (one of the Treg cytokines) decreased, but there was no significant difference in IL-17A. Silencing the miR-106b-5p of CD4+ T-cells can promote its polarization to Treg. After up-regulating the expression of miR-106b-5p on CD4+ T cells, the expressions of NR4A3, Foxp3, and TGF-beta all decreased. These results were reversed by NR4A3, but IL-17A remained unchanged. Inhibition of miR-106b-5p expression in the experimental ITP model resulted in an increase in peripheral blood platelet count and spleen Treg frequency but no change in Th17 frequency in ITP model mice.
MiR-125a-5p targeted CXCL13 participates in the pathological process of ITP and is regulated by long non-coding RNA MEG3 to induce immune imbalance of Treg/Th17 in ITP.77,152 The authors observed that the concentration of plasma CXCL13 in PBMCs of patients with ITP increased and negatively correlated with miR-125a-5p. After CD4+ T-cells up-regulated/down-regulated miR-125a-5p, CXCL13 decreased/increased. Dexamethasone, a first-line treatment for ITP, upregulated miR-125a-5p expression and decreased CXCL13 in CD4+ T-cells, and miR-125a-5p inhibitors could reverse this process. It is suggested that CXCL13 is the target of miR-125a-5p and participates in the pathological process of ITP, which is consistent with the results of previous studies. 149 Subsequent studies have shown that MEG3, a long non-coding RNA highly expressed in CD4+ T-cells of ITP patients, directly interacts with miR-125a-5p and inhibits its expression. The expression of MEG3 in CD4+ T-cells decreased after being treated with dexamethasone. Down-regulation of MEG3 or overexpression of miR-125a-5p in CD4+ T-cells can promote Treg polarization and inhibit Th17 polarization.
Two miRNAs were verified to be involved in the pathogenesis of ITP through animal experiments. The expression of miRNA-30a in splenic mononuclear cells of the ITP mouse model is increased, which is positively correlated with the expression of ROR γt. It is involved in the pathogenesis of ITP by affecting the differentiation of Th17 cells. The authors used biological software to predict that miRNA-30a may target SOCS3, a cytokine signal inhibitor that inhibits the differentiation of Th17 cells. Unfortunately, further verification shows that although SOCS3 can bind to the target site of miRNA-30a, it may not be its functional target gene. 78 The latest study found that miR-199a-5p participates in the pathological process of ITP by inhibiting the differentiation of Th17. 79 The level of miR-199a-5p in the spleen CD4+ T cells of the ITP mouse model decreased, while the protein levels of STAT3 and pSTAT3 increased. MiR-199a-5p overexpression inhibited the differentiation of Th17 cells by targeting STAT3. The authors also developed an extracellular vesicle containing miR-199a-5p, which comes from adipose-derived mesenchymal stem cells modified by mir-199a-5p, and can inhibit Th17 differentiation in vitro and in vivo, increase platelet count in ITP mice, and have potential therapeutic effects on ITP.
Mesenchymal Stem Cells Interfere with the Platelet Growth Environment
Mesenchymal stem cells (MSCs) are pluripotent progenitor cells with the ability to differentiate into different cell lines, which can be obtained from bone marrow, umbilical cord blood, fat, dental pulp, and other tissues. 153 MSCs can regulate immune cells in both acquired immunity and innate immunity, including inhibiting the activation of B and T cells, inducing the production of tolerant dendritic cells, inducing macrophages to M2 polarization, and reducing the cytotoxicity of NK and NKT cells.153,154 There are quantitative dysfunction and immunomodulatory deficiency in bone marrow MSCs of patients with ITP, which lead to abnormal megakaryocyte development, disturbance of platelet production and immune intolerance, which may be related to Notch-1/Jagge-1 and TNFAIP3/NF-κB/SMAD7. 155 All-trans retinoic acid, Platelet-derived growth factor, and Thalidomide can partially correct the abnormsality of bone marrow MSCs in ITP.156,157 Due to the immunomodulatory effect of MSCs, MSCs transplantation has shown a certain therapeutic effect on ITP in vivo and in vitro, and in the clinic.158–161
Interestingly, MSC-derived extracellular vesicles (MSC-EV) have the same effect and have higher safety and lower immunogenicity. Using MSC-EV as a substitute for MSC can avoid complications such as ectopic tumor formation and immune rejection, and has many advantages. 162 Li et al 79 developed extracellular vesicles containing miR-199a-5p, which is derived from mir-199a-5p modified adipose-derived mesenchymal stem cells. It was found that the miR-199a-5p level of CD4+ T-cells in the spleen of ITP mice decreased, while the protein levels of STAT3 and pSTAT3 (Th17 transcription factor) increased. The overexpression of miR-199a-5p inhibited the differentiation of Th17 cells by targeting STAT3. Adipose MSC-EV-derived miR-199a-5p can inhibit Th17 differentiation in vitro and in vivo, restore Th17/Treg balance, increase platelet count in ITP mice, and have potential therapeutic effects on ITP. Similarly, the expression of miR-146a-5p in bone marrow MSC-derived EVs is higher than that in bone marrow MSC, which can be directly related to IL-1R-associated kinase (IRAK1), inhibit its expression in CD4+ T-cells, and thus restore the Th17/Treg ratio. 69 In the previous study, 62 miRNAs with a significant difference in bone marrow MSC between ITP patients and healthy controls were detected, of which 5 (miR-98-5p, miR-20b-5p, miR-3148, miR-4284 miR-3977) increased continuously in ITP-MSCs after some of them were verified by qPCR. 163 The researchers were targeting miR-98-5p because it had the highest fold change in micro-array analysis and because of its relevance to cell apoptosis. It was found that after up-regulation of miR-98-5p in MSCs from healthy volunteers, the state of cells was similar to that of MSCs in ITP patients, showing abnormal morphology and increased apoptosis rate, while inhibition of miR-98-5p expression in MSCs derived from ITP patients improved cell morphology and increased survival rate. Overexpression of miR-98-5p in MSCs weakens the therapeutic effect of MSCs on ITP mice. MiR-98-5p acts by targeting inhibition of insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1). The subsequent down-regulation of insulin-like growth factor 2 (IGF-2) leads to the PI3K/Akt pathway and participates in the process of MSC deficiency. In addition, miR-98-5p upregulates p53-TrCP-dependent p53 ubiquitination by inhibiting the ubiquitination of p53 containing β-transduction protein repeat sequences, which leads to the accumulation of p53 and down-regulation of IGF1R (an IGF2 receptor), which also leads to the inhibition of PI3K/AKt pathway, resulting in MSC deficiency. PI3K/AKt pathway and β-TrCP/p53 ubiquitin pathway cross-affect the apoptosis of ITP-MSCs, which is related to the targeted inhibition of IGF2BP1 by miR-98-5p. In addition, the authors found that all-trans retinoic acid (ATRA) protects MSCs from apoptosis by down-regulating miR-98-5p, thus providing a potential treatment for ITP. 80
Megakaryocytes and Platelets
Megakaryocytes are highly specialized hematopoietic progenitor cells, which are responsible for the production of platelets in the bone marrow and may be the target of ITP immune-mediated injury. There are some contradictory views on megakaryocytes in ITP. Some studies have shown that ITP megakaryocytes show ultrastructural characteristics of apoptosis, while other studies have shown that ITP megakaryocytes are resistant to apoptosis. 164 Abnormal autophagy of megakaryocytes in patients with ITP affects the differentiation of hematopoietic stem cells into megakaryocytes and platelet production by megakaryocytes. 165 Anti-platelet antibodies can impair the ability of megakaryocytes to release platelets. 10 Abnormal megakaryocytes and thrombocytopenia are caused by a variety of reasons in patients with ITP. The important role of miRNA in megakaryocyte and platelet production has been determined.166,167
Considering that some patients with acute ITP or acute exacerbation of chronic ITP have a history of viral infection from days to weeks before the onset of thrombocytopenia and that viral miRNA can regulate viral gene expression or host cell gene expression, the authors studied the expression of viral miRNA in ITP. 81 Umbilical cord blood-derived mononuclear cells were cultured with plasma from ITP patients and healthy people to incubate megakaryocytes. The plasma of patients with ITP significantly inhibited the number and maturity of megakaryocytes. Gene chip analysis showed that 14 virus miRNA were up-regulated in the experimental group (hsv1-miR-H7* hsv2-miR-H9-3p ebv-miR-BART19-3p hsv2-miR-H24 hsv2-miR-H10 hsv1-miR-H17 hsv1-miR-H8* ebv-miR-BART6-3p hsv1-miR-H14-3p ebv-miR-BART8* sv40-miR-S1-5p ebv-miR-BHRF1-2 hsv2-miR-H7-3p hsv2-miR-H6). Bioinformatics analysis shows that they may interfere with megakaryocyte maturation by regulating genes related to cell development, metabolic process and, cell biosynthesis. Further studies showed that EBV-miR-BART6 down-regulated the expression of miR-185-5p in megakaryocytes, which triggered the upregulation of ABCG4 gene expression. 168 This effect is accompanied by the decrease of megakaryocyte maturation, development, differentiation, and cell proliferation.
EBV-miR-BART6-5p may target Dicer1 to promote proliferation and metastasis of nasopharyngeal carcinoma cells and inhibit apoptosis.169,170 However, miR-BART6-3p encoded by EBV can inhibit tumors. It can target and down-regulate long non-coding RNA LOC553103 and affect the stability of 3′UTR of STMN1 mRNA, so as to regulate the expression of cell cycle-related proteins and inhibit the proliferation of EBV-related tumor cells. 171 According to the “asymmetry rule”, the miR-BART6 double chain predicts that the 3p chain is selected as the effective chain because the stability of the 5′ end of the 3p chain is worse. However, it was noted that the more stable 5p chain at the 5′ end of the double chain is more effectively loaded into miRISC as the effective chain.46,169 EBV miR-BART6-5p combined with dexitabine can enhance the expression of miR-29 in EBV-positive Burkitt lymphoma cell lines. 48 MiR-BART6-3p avoids RIG-I recognition by targeting RIG-I (a pattern recognition receptor), evades the host's innate immune response, and promotes EBV infection. 172
MiR-557 is highly expressed in human megakaryocytic leukemia cells and plasma of children with ITP, its inhibitors improve ITP by regulating apoptosis-related genes and cellular immunity and activating Akt/ERK pathway. 82 MiR-557 inhibitors may significantly inhibit the excessive apoptosis of megakaryocytes by reducing the expression of apoptosis genes caspase-3 and Bax. MiR-557 inhibitors can not only increase the cell survival rate, inhibit apoptosis and improve the symptoms of hemorrhage in ITP model rats, but also increase the number of platelets and megakaryocytes, promote TPO-induced megakaryocyte multinucleation, and promote cell division. In addition, miR-577 inhibitors could increase the ratio of Treg/CD4+ T-cells, increase the level of TGF-beta and decrease the level of IL-6 in peripheral blood of rats.
Platelets are essential for proper hemostasis and thrombosis. Although platelets lack nuclei, there are functional splicing bodies and signal-dependent precursor mRNA splicing in them, as well as miRNA and the processing mechanism of miRNA.173,174 Although the number of miRNA in platelets is significantly lower than that in nucleated cells, it is also rich in species, accounting for about 30% of the currently annotated mature human miRNA. 175 There is evidence that although a large proportion of platelet miRNA content comes from megakaryocytes, platelets can also absorb miRNA from the environment and transfer it to other cells.176,177 Among them, miR-96 regulates the expression of vesicle associated membrane protein 8 (also known as endobrevin), while miR-223 plays an opposite role in platelet production and function.174,178 In recent years, the research on the target of miRNA in ITP is becoming more and more clear.
Microarray analysis was used to compare the expression of miRNA in platelets of patients with ITP and healthy controls. There were 115 miRNAs differentially expressed between the two groups. 83 Nine miRNAs were further verified by RT-qPCR, and the results showed that 4 miRNAs were only expressed in ITP (miR-338-5p, miR-765, miR-122-5p and miR-451b), and the other 5 miRNAs were significantly decreased in ITP (miR-133a-3p,miR-224-3p miRNA miR-452-5p, miR-491-5p and miR-15a-3p). Bioinformatics analysis of 115 differentially expressed miRNAs showed that miR-548a-5p was the most important regulator of down-regulated miRNA and miR-6867-5p was the most important regulator of up-regulated miRNA; they regulated 50 and 24 related target genes respectively. Multiple miRNAs may jointly regulate celladhesionmolecules (CAMs) pathway and play an important role in platelet cell adhesion, which may explain the role of CAMs pathway in the pathogenesis of ITP. 179
Circulating Cell-Free miRNA
In addition to nucleated cells and platelets, miRNA also exists in the extracellular environment, which is wrapped in extracellular vesicles and circulates with body fluids to establish communication between cells. 180 Circulating cell-free miRNA comes from different types of cells and exists in plasma or serum, urine, saliva, and other body fluids, and the content is stable. As a non-invasive “fluid biopsy”, cell-free miRNA analysis provides a promising strategy for disease diagnosis, prognosis, and monitoring. For example, specific cell-free miRNA shows good diagnostic effectiveness in many diseases, such as cancer, central nervous system diseases, obesity and so on.181,182 There is no doubt about the change of miRNA in plasma or serum of ITP patients (Appendix A), but it is not clear to what extent the circulating miRNA spectrum of patients with ITP has changed, and whether the spectrum of this change can be used as a biomarker to improve the diagnostic level of ITP.
Two studies used Agilent human miRNA microarray (Version 19) to analyze plasma miRNA profiles. Unfortunately, miR-4499 up-regulated, which was the only similarity between the two studies, although both studies used qRCP to verify part of the miRNA to confirm the accuracy of the microarray results.183,184 The main reason for this result may be the difference in the number of included cases, This will require further study.
In ROC analysis, 10 miRNAs (miR-144-3p, miR-320c, miR-642b-3p, miR-1275, miR-3141, miR-3162-3p, miR-4270, miR-4499, miR-4739 and miR-6126) had positive diagnostic value when tested alone. 184 Of those 10 miRNAs, miR-3141 had the best AUC value of 0.729 (95% CI: 0.627 to 0.830). When the four (miR-144-3p, miR-1275, miR-3141 and miR-3162-3p) miRNAs were analyzed as a group or together with antiplatelet autoantibodies, the diagnostic value was increased. It was also found that the level of plasma miR-3162-3p was positively correlated with platelet count in patients with ITP.
As the disease progresses, so does miRNA. For example, the expression of miR-642-3p,miR-1275,miR-3141, and miR-4499 is up-regulated in newly diagnosed ITP patients, while plasma miR-144-3p,miR-320C,miR-3162-3p, and miR-6126 levels are changed in patients with persistent and chronic ITP. Plasma miR-320c levels were significantly increased in ITP patients who responded to hormone therapy. 184 In the plasma of children with ITP, the level of miR-302c-3p in patients with acute ITP was 4.4 times higher than that in patients with chronic ITP (P < 0.028). 185 As for miR-223-3p, acute ITP is 27.59 times higher than that of chronic ITP patients.
Another study isolated exosomes from plasma of patients with ITP for proteomic analysis and RNA sequencing, and found 16 differentially expressed miRNA (6 down-regulated and 10 up-regulated (Supplemental Figures A), and one up-regulated exosome protein (apolipoprotein E, ApoE). The expression of three exosomes miRNA was evaluated by qPCR. In ITP patients, the expression of miR-142-5p and miR-29b-3p increased, while the expression of miR-584-5p decreased. 186
There were 81 miRNAs differentially expressed in ITP patients. Using droplet digital PCR for verification, three kinds of miRNA differential expression were found in ITP patients before TPO-RA treatment: miR-199a-5p down-regulated, miR-33a-5p, and miR-195-5p up-regulated. ROC curve analysis showed that the combination of miR-199a-5p and miR-33a-5p could distinguish ITP patients from the control group, and the AUC was 0.93. 187
The change of plasma miRNA level may reflect the potential autoimmune response of ITP and subsequent platelet destruction. The source of plasma miRNA cells is still unknown, which may come from damaged or apoptotic cells or platelets, or from changes in an autoimmune response. The results of the above independent studies are very different. There is still a long way to go for circulating cell-free miRNA to be used as a “fluid biopsy” to guide the prognosis of ITP, which may be the focus of research in the future.
Differential Expression of miRNA Before and After Medical Treatment
The aim of ITP treatment is to raise the platelet count to a safe level to reduce the risk of death from massive bleeding. The standard of care for newly diagnosed patients remains corticosteroid.188–190 TPO receptor agonist (TPO-RA) is a safe and effective treatment option for chronic ITP patients who are at risk of bleeding after first-line treatment failure. The continuous response to TPO-RA can last as long as 6 to 8 years. 189 Thrombopoietin (TPO), synthesized in the liver, induces intracellular signaling cascades by binding and activating TPO receptors on the surface of megakaryocytes. 191
Using miRNA PCR panel analysis, 14 miRNAs showed significant changes during TPO-RA treatment. Quantitative analysis by ddPCR showed that 6 miRNAs (miR-199a-5p, miR-33a-5p, miR-382-5p, miR-92b-3p, miR-26a-5p and miR-221-3p) changed after treatment with TPO-RAs, of which 5 (except miR-33a-5p) increased 2 weeks after TPO-RAS. MiR-33a-5p decreased 6 weeks after TPO-RAs compared with that before treatment. The level of miR-195-5p was not affected by TPO-RAs and was significantly higher than that in the control group, which may indicate that it may play a role in the pathophysiology of ITP. The levels of miR-199a-5p and miR-221-3p before treatment are helpful to predict the response of platelets to TPO-RA treatment. 187
The expression of 25 miRNAs (Supplemental Figures A) was down-regulated in ITP patients (p < 0.005), and the expression level was similar to that of the normal control group after QSBLE treatment. 44 miRNAs were differentially expressed between the treatment group and the ITP group (35 up-regulated and 9 down-regulated), of which 14 targeted 31 known ITP-related genes. 13 of the 14 miRNA were up-regulated after QSBLE treatment. Bioinformatics analysis showed that most of the 31 known ITP related genes were involved in the immune process of B cells and T cells. the treatment of ITP by QSBLE may be achieved by up-regulating and restoring part of miRNA to suppress many important immunoreactive genes. 91
Conculsion
In the past few years, the research on miRNA has made rapid progress, especially in immunology (Figure 2). This provides new insights into the pathophysiology of ITP. The relationship between miRNA action mechanism and ITP is complex, and we only understand a small part of it. In this review, we focus on some of the detailed changes known in immunology. The results of many studies point to the abnormal expression of miRNAs such as miR-155, miR-146, mir-142 and miR-181, which play an important role in the pathophysiology of ITP. This also shows that they are hotspots in current research. The detection of ITP related miRNA can be used as a correct potential biomarker for the diagnosis and prognosis of ITP. The levels of miR-199a-5p and miR-221-3p are helpful to predict the response of patients to TPO-RAs treatment. The level of miR-155 is helpful to predict the prognosis of elderly patients. It is worth noting that most of the results described have not been reproduced, which may be due to blood sources, different population sizes and different races studied, as well as the use of different RNA isolation and detection technologies. Therefore, standardized detection schemes are needed in the future, including more sensitive miRNA detection methods and quantitative analysis models. In conclusion, it may be very promising to continue to analyze the role of miRNA in the development and progress of ITP.
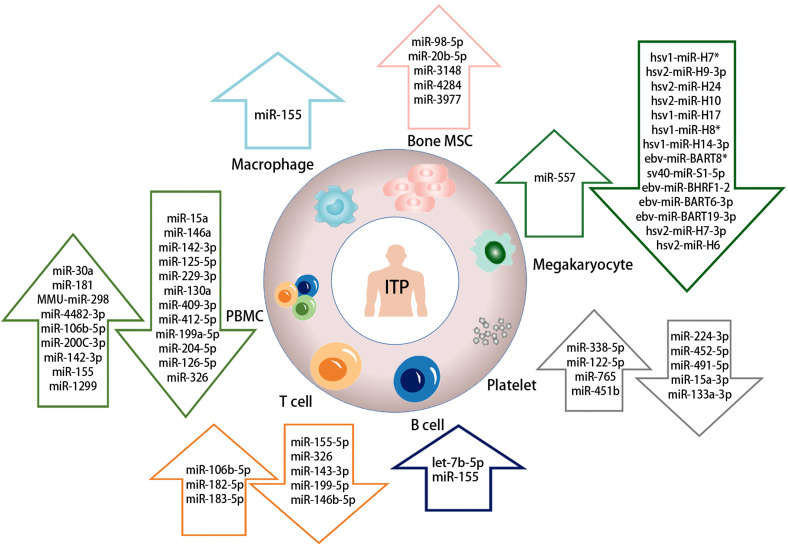
Figure 2.
Abnormal expression of cell-associated miRNAs in ITP.
Supplementary Material
Appendix A
Differential Expression of Circulating Cell-Free MiRNA in ITP
| Upregulated | Downregulated | ||
|---|---|---|---|
| miR-2234-5p | miR-5787 | miR-26-5p | miR-130b-5p |
| miR-4433-3p | miR-320c | miR-244-3p | miR-222-3p |
| miR-3242 | miR-6126 | miR-206b-5p | miR-374b-5p |
| miR-4499 | miR-302c-3p | miR-25a-5p | miR-107 |
| miR-234 | miR-223-3p | miR-202-3p | miR-766-3p |
| miR-2392 | miR-597 | miR-4707-5p | miR-191-5p |
| miR-4778 -5p | miR-29a-3p | miR-4722 | miR-339-5p |
| miR-483-5p | miR-142-5p | miR-2290 | miR-223-3p |
| miR-4800-5p | miR -29b-3p | miR-297-3p | miR-199a-5p |
| miR-3667-5p | miR-16-2-3p | miR-3620-3p | miR-103a-3p |
| miR-630 | miR-501-3p | miR-378i | miR-30b-5p |
| miR-5703 | miR-144-5p | miR-4454 | |
| miR-652-5p | miR-192-5p | miR-342-3p | |
| miR-765 | miR-182-5p | miR-498 | |
| miR-3202 | miR-363-3p | miR-3162-3p | |
| miR-4739 | miR-96-5p | miR-144-3p | |
| miR-5195-3p | miR-1234-5p | miR-1281 | |
| miR-4270 | miR-642b-3p | miR-302a-3p | |
| miR-1275 | miR-1234-5p | miR-544 | |
| miR-3141 | miR-557 | miR-584-5p | |
| miR-486-5p | miR-1260a | miR-4433a-5p | |
| miR-629-5p | miR-423-5p | miR- 4433b-3p | |
| miR-222-3p | miR-378a-3p | miR-6842-3p | |
Footnotes
Authors’ Contribution: Conceptualization, RR.X. and YR.Z; the literature search and summary, SY.C. and Y.W.; writing-original draft, YR.Z.; review—writing and editing, YR.Z. and RR.X. All authors have read and agreed to the published version of the manuscript.
Authors’ Note: Both SY.C. and Y.W. wrote the manuscript equally.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the the Natural Science Foundation of Shandong Province, (grant number ZR2020KH023).
ORCID iD: Ruirong Xu https://orcid.org/0000-0002-9871-1542
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Piel-Julian ML, Mahévas M, Germain J, et al. Risk factors for bleeding, including platelet count threshold, in newly diagnosed immune thrombocytopenia adults. J Thromb Haemost. 2018;16(9):1830-1842. [DOI] [PubMed] [Google Scholar]
- 2.Efficace F, Mandelli F, Fazi P, et al. Health-related quality of life and burden of fatigue in patients with primary immune thrombocytopenia by phase of disease. Am J Hematol. 2016;91(10):995-1001. [DOI] [PubMed] [Google Scholar]
- 3.Schoonen WM, Kucera G, Coalson J, et al. Epidemiology of immune thrombocytopenic purpura in the general practice research database. Br J Haematol. 2009;145(2):235-244. [DOI] [PubMed] [Google Scholar]
- 4.Moulis G, Palmaro A, Montastruc JL, Godeau B, Lapeyre-Mestre M, Sailler L. Epidemiology of incident immune thrombocytopenia: a nationwide population-based study in France. Blood. 2014;124(22):3308-3315. [DOI] [PubMed] [Google Scholar]
- 5.He R, Reid DM, Jones CE, Shulman NR. Spectrum of Ig classes, specificities, and titers of serum antiglycoproteins in chronic idiopathic thrombocytopenic purpura. Blood. 1994;83(4):1024-1032. [PubMed] [Google Scholar]
- 6.Saleh MN, Moore DL, Lee JY, LoBuglio AF. Monocyte-platelet interaction in immune and nonimmune thrombocytopenia. Blood. 1989;74(4):1328-1331. [PubMed] [Google Scholar]
- 7.Li J, van der Wal DE, Zhu G, et al. Desialylation is a mechanism of Fc-independent platelet clearance and a therapeutic target in immune thrombocytopenia. Nat Commun. 2015;6:7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrbensky JR, Nazy I, Toltl LJ, et al. Megakaryocyte apoptosis in immune thrombocytopenia. Platelets. 2018;29(7):729-732. [DOI] [PubMed] [Google Scholar]
- 9.Grodzielski M, Di Buduo CA, Goette NP, et al. Autoantibodies in immune thrombocytopenia affect the physiological interaction between megakaryocytes and bone marrow extracellular matrix proteins. Br J Haematol. 2018;183(2):319-323. [DOI] [PubMed] [Google Scholar]
- 10.Iraqi M, Perdomo J, Yan F, Choi PY, Chong BH. Immune thrombocytopenia: antiplatelet autoantibodies inhibit proplatelet formation by megakaryocytes and impair platelet production in vitro. Haematologica. 2015;100(5):623-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsson B, Andersson PO, Jernås M, et al. T-cell-mediated cytotoxicity toward platelets in chronic idiopathic thrombocytopenic purpura. Nat Med. 2003;9(9):1123-1124. [DOI] [PubMed] [Google Scholar]
- 12.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9-14. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pu M, Chen J, Tao Z, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76(3):441-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843-854. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21(17):4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libri V, Miesen P, van Rij RP, Buck AH. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell Mol Life Sci. 2013;70(19):3525-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozsolak F, Poling LL, Wang Z, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22(22):3172-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monteys AM, Spengler RM, Wan J, et al. Structure and activity of putative intronic miRNA promoters. Rna. 2010;16(3):495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23(20):4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moss EG. MicroRNAs: hidden in the genome. Curr Biol. 2002;12(4):R138-R140. [DOI] [PubMed] [Google Scholar]
- 23.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432(7014):231-235. [DOI] [PubMed] [Google Scholar]
- 24.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. Melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14(23):2162-2167. [DOI] [PubMed] [Google Scholar]
- 25.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235-240. [DOI] [PubMed] [Google Scholar]
- 26.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14(4):156-159. [DOI] [PubMed] [Google Scholar]
- 28.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95-98. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y, Yeo J, Lee JH, et al. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014;9(3):1061-1074. [DOI] [PubMed] [Google Scholar]
- 30.Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57(3):397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwasaki S, Kobayashi M, Yoda M, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39(2):292-299. [DOI] [PubMed] [Google Scholar]
- 32.Pare JM, Tahbaz N, López-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20(14):3273-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwasaki S, Sasaki HM, Sakaguchi Y, Suzuki T, Tadakuma H, Tomari Y. Defining fundamental steps in the assembly of the drosophila RNAi enzyme complex. Nature. 2015;521(7553):533-536. [DOI] [PubMed] [Google Scholar]
- 34.Ye X, Huang N, Liu Y, et al. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18(6):650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Ye X, Jiang F, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325(5941):750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437-1441. [DOI] [PubMed] [Google Scholar]
- 37.Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130(2):287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin C. Cleavage of the star strand facilitates assembly of some microRNAs into Ago2-containing silencing complexes in mammals. Mol Cells. 2008;26(3):308-313. [PubMed] [Google Scholar]
- 39.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185-197. [DOI] [PubMed] [Google Scholar]
- 40.Park JH, Shin C. Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res. 2015;43(19):9418-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16(9):953-960. [DOI] [PubMed] [Google Scholar]
- 42.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. Rna. 2010;16(1):43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomari Y, Du T, Zamore PD. Sorting of drosophila small silencing RNAs. Cell. 2007;130(2):299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoda M, Kawamata T, Paroo Z, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17(1):17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130(1):101-112. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199-208. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki HI, Katsura A, Yasuda T, et al. Small-RNA asymmetry is directly driven by mammalian argonautes. Nat Struct Mol Biol. 2015;22(7):512-521. [DOI] [PubMed] [Google Scholar]
- 48.Mazzoccoli L, Robaina MC, Bacchi CE, Soares LS, Klumb CE. Mir-29 promoter and enhancer methylation identified by pyrosequencing in Burkitt lymhoma cells: interplay between MYC and miR-29 regulation. Oncol Rep. 2019;42(2):775-784. [DOI] [PubMed] [Google Scholar]
- 49.Beilharz TH, Humphreys DT, Clancy JL, et al. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. Plos One. 2009;4(8):e6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103(11):4034-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin F, Hu H, Xu M, et al. Serum microRNA profiles serve as novel biomarkers for autoimmune diseases. Front Immunol. 2018;9:2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, Xu X, Yang J. miRNAs Alter T helper 17 cell fate in the pathogenesis of autoimmune diseases. Front Immunol. 2021;12:593473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Q, Chu L, Li Y, et al. miR-23a/b suppress cGAS-mediated innate and autoimmunity. Cell Mol Immunol. 2021;18(5):1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Intern Med. 2015;278(4):369-395. [DOI] [PubMed] [Google Scholar]
- 56.Hammad MHR, Hamed D, Eldosoky M, et al. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 2018;24(3):171-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khoshmirsafa M, Kianmehr N, Falak R, et al. Elevated expression of miR-21 and miR-155 in peripheral blood mononuclear cells as potential biomarkers for lupus nephritis. Int J Rheum Dis. 2019;22(3):458-467. [DOI] [PubMed] [Google Scholar]
- 58.Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev. 2011;27(8):862-866. [DOI] [PubMed] [Google Scholar]
- 59.Honardoost MA, Kiani-Esfahani A, Ghaedi K, Etemadifar M, Salehi M. miR-326 and miR-26a, two potential markers for diagnosis of relapse and remission phases in patient with relapsing-remitting multiple sclerosis. Gene. 2014;544(2):128-133. [DOI] [PubMed] [Google Scholar]
- 60.Chang Y, Chen X, Tian Y, et al. Downregulation of microRNA-155–5p prevents immune thrombocytopenia by promoting macrophage M2 polarization via the SOCS1-dependent PD1/PDL1 pathway. Life Sci. 2020;257:118057. [DOI] [PubMed] [Google Scholar]
- 61.Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001-1009. [DOI] [PubMed] [Google Scholar]
- 62.Zhao H, Li H, Du W, et al. Reduced MIR130A is involved in primary immune thrombocytopenia via targeting TGFB1 and IL18. Br J Haematol. 2014;166(5):767-773. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Zhao H, Xue F, et al. Reduced expression of MIR409-3p in primary immune thrombocytopenia. Br J Haematol. 2013;161(1):128-135. [DOI] [PubMed] [Google Scholar]
- 64.Qian C, Yan W, Li T, et al. Differential expression of MiR-106b-5p and MiR-200c-3p in newly diagnosed versus chronic primary immune thrombocytopenia patients based on systematic analysis. Cell Physiol Biochem. 2018;45(1):301-318. [DOI] [PubMed] [Google Scholar]
- 65.Li JQ, Tian JM, Fan XR, et al. miR-106b-5p induces immune imbalance of Treg/Th17 in immune thrombocytopenic purpura through NR4A3/Foxp3 pathway. Cell Cycle. 2020;19(11):1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Y, Zhu H, Xie X, Zheng Z, Ling Y. MicroRNA expression profile in Treg cells in the course of primary immune thrombocytopenia. J Investig Med. 2019;67(8):1118-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu L, Hua M, Liu C, He N, Li Z, Ma D. The aberrant expression of microRNAs and correlations with T cell subsets in patients with immune thrombocytopenia. Oncotarget. 2016;7(47):76453-76463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hua M, Li J, Wang C, et al. Aberrant expression of microRNA in CD4(+) cells contributes to Th17/Treg imbalance in primary immune thrombocytopenia. Thromb Res. 2019;177:70-78. [DOI] [PubMed] [Google Scholar]
- 69.He Y, Ji D, Lu W, et al. Bone marrow mesenchymal stem cell-derived exosomes induce the Th17/Treg imbalance in immune thrombocytopenia through miR-146a-5p/IRAK1 axis. Hum Cell. 2021;34(5):1360-1374. [DOI] [PubMed] [Google Scholar]
- 70.He YZ, Lu RF, Zhu C, Hua JY. Qian five Rhinoceros Gindeng (QFRG) protects against development of immune thrombocytopenia via miR-181a inhibition of TLR-4 expression. Int J Clin Exp Med. 2015;8(5):6986-6993. [PMC free article] [PubMed] [Google Scholar]
- 71.Chang Y, Chen X, Tian Y, et al. Downregulation of microRNA-155-5p prevents immune thrombocytopenia by promoting macrophage M2 polarization via the SOCS1-dependent PD1/PDL1 pathway. Life Sci. 2020;257:118057. [DOI] [PubMed] [Google Scholar]
- 72.He Y, Wei XH, Zao XM. [Value of MiR-155 in prognostic evaluation of elderly patients with primary immune thrombocytopenia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(4):1338-1343. [DOI] [PubMed] [Google Scholar]
- 73.Qian BH, Ye X, Zhang L, et al. Increased miR-155 expression in peripheral blood mononuclear cells of primary immune thrombocytopenia patients was correlated with serum cytokine profiles. Acta Haematol. 2015;133(3):257-263. [DOI] [PubMed] [Google Scholar]
- 74.Guo Y, Qu W, Wang YH, et al. [The role of miR-155 in pathogenesis of immune thrombocytopenia]. Zhonghua Yi Xue Za Zhi. 2016;96(14):1103-1107. [DOI] [PubMed] [Google Scholar]
- 75.Feng Y, Geng LL, Li XQ, Nan N. [Expression of MicroRNA-15a in children with primary immune thrombocytopenia and its significance]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017;25(6):1772-1775. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Jia X, Zhou L, et al. Increased let-7b-5p is associated with enhanced BAFF-R expression and B cell survival in immune thrombocytopenia. Int Immunopharmacol. 2021;93:107393. [DOI] [PubMed] [Google Scholar]
- 77.Li JQ, Hu SY, Wang ZY, et al. MicroRNA-125-5p targeted CXCL13: a potential biomarker associated with immune thrombocytopenia. Am J Transl Res. 2015;7(4):772-780. [PMC free article] [PubMed] [Google Scholar]
- 78.Wu XF, Li JQ, Hu SY, et al. [Pathogenesis of immune thrombocytopenic Purpura (ITP) by MiRNA-30a-mediated Th17 cell differentiation]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(2):588-594. [DOI] [PubMed] [Google Scholar]
- 79.Li J, Xia Y, Fan X, et al. Extracellular vesicles derived from miR-199a-5p-modified adipose-derived mesenchymal stem cells alleviate immune thrombocytopenia by inhibiting T helper 17 differentiation. Lab Invest. 2021;101(3):318-327. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Zhang J, Su Y, et al. miRNA-98-5p targeting IGF2BP1 induces mesenchymal stem cell apoptosis by modulating PI3K/Akt and p53 in immune thrombocytopenia. Mol Ther Nucleic Acids. 2020;20:764-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, Huang R, Song C, Hu H, Zhang M. Some viral microRNAs were up-regulated in megakaryocytes incubated with immune thrombocytopenia plasma. Eur J Haematol. 2013;90(3):220-227. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Guo Y, Zhang X, et al. The role and mechanism of miR-557 in inhibiting the differentiation and maturation of megakaryocytes in immune thrombocytopenia. Rna Biol. 2021;18(11):1953-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deng G, Yu S, He Y, et al. MicroRNA profiling of platelets from immune thrombocytopenia and target gene prediction. Mol Med Rep. 2017;16(3):2835-2843. [DOI] [PubMed] [Google Scholar]
- 84.The Impact of Food Bioactives on Health: In Vitro and ex Vivo Models. The Impact of Food Bioactives on Health: In Vitro and ex Vivo Models. Springer; 2015. [PubMed] [Google Scholar]
- 85.Dai Y, Huang YS, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16(12):939-946. [DOI] [PubMed] [Google Scholar]
- 86.Qiao J, Liu Y, Wu Y, et al. Aberrant expression of RUNX3 in patients with immune thrombocytopenia. Int Immunopharmacol. 2015;28(1):252-256. [DOI] [PubMed] [Google Scholar]
- 87.Lu L, Gasteiger G, Yu I, et al. A single miRNA-mRNA interaction affects the immune response in a context- and cell-type-specific manner. Immunity. 2015;43(1):52-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu C, Huang X, Zhang X, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117(16):4293-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang K, Yang J, Guo S, Zhao G, Wu H, Deng G. Peripheral circulating exosome-mediated delivery of miR-155 as a novel mechanism for acute lung inflammation. Mol Ther. 2019;27(10):1758-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou X, Yan T, Huang C, et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J Exp Clin Cancer Res. 2018;37(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burenbatu, Borjigin M, Eerdunduleng, et al. Profiling of miRNA expression in immune thrombocytopenia patients before and after Qishunbaolier (QSBLE) treatment. Biomed Pharmacother. 2015;75:196-204. [DOI] [PubMed] [Google Scholar]
- 92.Doyle AG, Herbein G, Montaner LJ, et al. Interleukin-13 alters the activation state of murine macrophages in vitro: comparison with interleukin-4 and interferon-gamma. Eur J Immunol. 1994;24(6):1441-1445. [DOI] [PubMed] [Google Scholar]
- 93.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176(1):287-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233(9):6425-6440. [DOI] [PubMed] [Google Scholar]
- 95.Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol. 2020;877:173090. [DOI] [PubMed] [Google Scholar]
- 96.Tavassoli M, McMillan R. Structure of the spleen in idiopathic thrombocytopenic purpura. Am J Clin Pathol. 1975;64(2):180-191. [DOI] [PubMed] [Google Scholar]
- 97.Audia S, Mahévas M, Samson M, Godeau B, Bonnotte B. Pathogenesis of immune thrombocytopenia. Autoimmun Rev. 2017;16(6):620-632. [DOI] [PubMed] [Google Scholar]
- 98.Essandoh K, Li Y, Huo J, Fan GC. MiRNA-Mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46(2):122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu HY, Yan Z, Xu Y, et al. Sequence features associated with microRNA strand selection in humans and flies. Bmc Genomics. 2009;10:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mycko MP, Cichalewska M, Cwiklinska H, Selmaj KW. miR-155-3p drives the development of autoimmune demyelination by regulation of heat shock protein 40. J Neurosci. 2015;35(50):16504-16515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu L, McCurdy S, Huang S, et al. Time series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci Rep. 2016;6:37446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Zhang M, Zhong M, Suo Q, Lv K. Expression profiles of miRNAs in polarized macrophages. Int J Mol Med. 2013;31(4):797-802. [DOI] [PubMed] [Google Scholar]
- 103.Wang G, Chen JJ, Deng WY, Ren K, Yin SH, Yu XH. CTRP12 Ameliorates atherosclerosis by promoting cholesterol efflux and inhibiting inflammatory response via the miR-155-5p/LXRα pathway. Cell Death Dis. 2021;12(3):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jost PJ, Ruland J. Aberrant NF-κB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood. 2007;109(7):2700-2707. [DOI] [PubMed] [Google Scholar]
- 105.Fu L, Linlee YC, Pham LV, Tamayo AT, Yoshimura LC, Ford RJ. BAFF-R promotes cell proliferation and survival through interaction with IKKbeta and NF-kappaB/c-Rel in the nucleus of normal and neoplastic B-lymphoid cells. Blood. 2009;113(19):4627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bi J, Zeng X, Zhao L, et al. miR-181a induces macrophage polarized to M2 phenotype and promotes M2 macrophage-mediated tumor cell metastasis by targeting KLF6 and C/EBPα. Mol Ther Nucleic Acids. 2016;5(9):e368. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 107.Siragam V, Crow AR, Brinc D, Song S, Freedman J, Lazarus AH. Intravenous immunoglobulin ameliorates ITP via activating fc gamma receptors on dendritic cells. Nat Med. 2006;12(6):688-692. [DOI] [PubMed] [Google Scholar]
- 108.Xu LL, Fu HX, Zhang JM, et al. Impaired function of bone marrow mesenchymal stem cells from immune thrombocytopenia patients in inducing regulatory dendritic cell differentiation through the notch-1/jagged-1 signaling pathway. Stem Cells Dev. 2017;26(22):1648-1661. [DOI] [PubMed] [Google Scholar]
- 109.El-Rashedi FH, El-Hawy MA, Helwa MA, Abd-Allah SS. Study of CD4(+), CD8(+), and natural killer cells (CD16(+), CD56(+)) in children with immune thrombocytopenic purpura. Hematol Oncol Stem Cell Ther. 2017;10(1):8-14. [DOI] [PubMed] [Google Scholar]
- 110.Zhang YJ, Qu W, Liu H, et al. [Quantities and function of NK cells in patients with immune thrombocytopenia]. Zhonghua Yi Xue Za Zhi. 2017;97(16):1231-1235. [DOI] [PubMed] [Google Scholar]
- 111.Harrington WJ, Minnich V, Hollingsworth JW, Moore CV. Demonstration of a thrombocytopenic factor in the blood of patients with thrombocytopenic purpura. J Lab Clin Med. 1951;38(1):1-10. [PubMed] [Google Scholar]
- 112.Karpatkin S, Siskind GW. In vitro detection of platelet antibody in patients with idiopathic thrombocytopenic purpura and systemic lupus erythematosus. Blood. 1969;33(6):795-812. [PubMed] [Google Scholar]
- 113.Roark JH, Bussel JB, Cines DB, Siegel DL. Genetic analysis of autoantibodies in idiopathic thrombocytopenic purpura reveals evidence of clonal expansion and somatic mutation. Blood. 2002;100(4):1388-1398. [PubMed] [Google Scholar]
- 114.van Leeuwen EF, van der Ven JT, Engelfriet CP, von dem Borne AE. Specificity of autoantibodies in autoimmune thrombocytopenia. Blood. 1982;59(1):23-26. [PubMed] [Google Scholar]
- 115.Neschadim A, Kotra LP, Branch DR. Small molecule phagocytosis inhibitors for immune cytopenias. Autoimmun Rev. 2016;15(8):843-847. [DOI] [PubMed] [Google Scholar]
- 116.Crow AR, Lazarus AH. Role of Fcgamma receptors in the pathogenesis and treatment of idiopathic. J Pediatr Hematol Oncol. 2003;25(Suppl 1):S14-S18. [DOI] [PubMed] [Google Scholar]
- 117.Yu TS, Wang HY, Zhao YJ, et al. Abnormalities of bone marrow B cells and plasma cells in primary immune thrombocytopenia. Blood Adv. 2021;5(20):4087-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Flint SM, Gibson A, Lucas G, et al. A distinct plasmablast and naïve B-cell phenotype in primary immune thrombocytopenia. Haematologica. 2016;101(6):698-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thien M, Phan TG, Gardam S, et al. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity. 2004;20(6):785-798. [DOI] [PubMed] [Google Scholar]
- 120.Sakai J, Akkoyunlu M. The role of BAFF system molecules in host response to pathogens. Clin Microbiol Rev. 2017;30(4):991-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stadanlick JE, Cancro MP. BAFF And the plasticity of peripheral B cell tolerance. Curr Opin Immunol. 2008;20(2):158-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hsu BL, Harless SM, Lindsley RC, Hilbert DM, Cancro MP. Cutting edge: BLyS enables survival of transitional and mature B cells through distinct mediators. J Immunol. 2002;168(12):5993-5996. [DOI] [PubMed] [Google Scholar]
- 123.Harless SM, Lentz VM, Sah AP, et al. Competition for BLyS-mediated signaling through Bcmd/BR3 regulates peripheral B lymphocyte numbers. Curr Biol. 2001;11(24):1986-1989. [DOI] [PubMed] [Google Scholar]
- 124.Zhu XJ, Shi Y, Peng J, et al. The effects of BAFF and BAFF-R-Fc fusion protein in immune thrombocytopenia. Blood. 2009;114(26):5362-5367. [DOI] [PubMed] [Google Scholar]
- 125.Zhou Z, Li X, Li J, et al. Direct B-cell stimulation by peripheral blood monocyte-derived dendritic cells in idiopathic thrombocytopenic purpura patients. J Clin Immunol. 2010;30(6):814-822. [DOI] [PubMed] [Google Scholar]
- 126.Ingold K, Zumsteg A, Tardivel A, et al. Identification of proteoglycans as the APRIL-specific binding partners. J Exp Med. 2005;201(9):1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Darce JR, Arendt BK, Wu X, Jelinek DF. Regulated expression of BAFF-binding receptors during human B cell differentiation. J Immunol. 2007;179(11):7276-7286. [DOI] [PubMed] [Google Scholar]
- 128.Yang Q, Xu S, Li X, et al. Pathway of toll-like receptor 7/B cell activating factor/B cell activating factor receptor plays a role in immune thrombocytopenia in vivo. Plos One. 2011;6(7):e22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Min YN, Wang CY, Li XX, et al. Participation of B-cell-activating factor receptors in the pathogenesis of immune thrombocytopenia. J Thromb Haemost. 2016;14(3):559-571. [DOI] [PubMed] [Google Scholar]
- 130.Akerlund J, Getahun A, Cambier JC. B cell expression of the SH2-containing inositol 5-phosphatase (SHIP-1) is required to establish anergy to high affinity, proteinacious autoantigens. J Autoimmun. 2015;62:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532(1):1-12. [DOI] [PubMed] [Google Scholar]
- 132.Pedersen IM, Otero D, Kao E, et al. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. Embo Mol Med. 2009;1(5):288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xue H, Hua LM, Guo M, Luo JM. SHIP1 Is targeted by miR-155 in acute myeloid leukemia. Oncol Rep. 2014;32(5):2253-2259. [DOI] [PubMed] [Google Scholar]
- 134.Lin X, Xu A, Zhou L, et al. Imbalance of T lymphocyte subsets in adult immune thrombocytopenia. Int J Gen Med. 2021;14:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kostic M, Zivkovic N, Cvetanovic A, Marjanović G. CD4(+) T cell phenotypes in the pathogenesis of immune thrombocytopenia. Cell Immunol. 2020;351:104096. [DOI] [PubMed] [Google Scholar]
- 136.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Legoux FP, Lim JB, Cauley AW, et al. CD4+ T Cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory T cells rather than deletion. Immunity. 2015;43(5):896-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Park H, Li Z, Yang XO, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6(11):1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Harrington LE, Hatton RD, Mangan PR, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123-1132. [DOI] [PubMed] [Google Scholar]
- 140.Carrier Y, Gao W, Korn T, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235-238. [DOI] [PubMed] [Google Scholar]
- 141.Lee GR. The balance of Th17 versus Treg cells in autoimmunity. Int J Mol Sci. 2018;19(3):730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485-517. [DOI] [PubMed] [Google Scholar]
- 143.WE P JZ. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238(1):247-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Paradowska-Gorycka A, Wajda A, Romanowska-Próchnicka K, et al. Th17/Treg-related transcriptional factor expression and cytokine profile in patients with rheumatoid arthritis. Front Immunol. 2020;11:572858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Britton GJ, Contijoch EJ, Mogno I, et al. Microbiotas from humans with inflammatory bowel disease Alter the balance of gut Th17 and RORγt(+) regulatory T cells and exacerbate colitis in mice. Immunity. 2019;50(1):212-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Talaat RM, Mohamed SF, Bassyouni IH, Raouf AA. Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: correlation with disease activity. Cytokine. 2015;72(2):146-153. [DOI] [PubMed] [Google Scholar]
- 147.Wang D, Huang S, Yuan X, et al. The regulation of the Treg/Th17 balance by mesenchymal stem cells in human systemic lupus erythematosus. Cell Mol Immunol. 2017;14(5):423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhong X, Wu Y, Liu Y, et al. Increased RUNX1 expression in patients with immune thrombocytopenia. Hum Immunol. 2016;77(8):687-691. [DOI] [PubMed] [Google Scholar]
- 149.Jernås M, Nookaew I, Wadenvik H, Olsson B. MicroRNA regulate immunological pathways in T-cells in immune thrombocytopenia (ITP). Blood. 2013;121(11):2095-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu R, Liu C, Chen D, et al. FOXP3 Controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015;75(8):1703-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zawada AM, Rogacev KS, Müller S, et al. Massive analysis of cDNA ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease. Epigenetics-Us. 2014;9(1):161-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li JQ, Hu SY, Wang ZY, et al. Long non-coding RNA MEG3 inhibits microRNA-125a-5p expression and induces immune imbalance of Treg/Th17 in immune thrombocytopenic purpura. Biomed Pharmacother. 2016;83:905-911. [DOI] [PubMed] [Google Scholar]
- 153.Fierabracci A, Del FA, Muraca M. The immunoregulatory activity of mesenchymal stem cells: ‘state of art’ and ‘future avenues’. Curr Med Chem. 2016;23(27):3014-3024. [DOI] [PubMed] [Google Scholar]
- 154.Yuan X, Qin X, Wang D, et al. Mesenchymal stem cell therapy induces FLT3L and CD1c( + ) dendritic cells in systemic lupus erythematosus patients. Nat Commun. 2019;10(1):2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.He Y, Xu LL, Feng FE, et al. Mesenchymal stem cell deficiency influences megakaryocytopoiesis through the TNFAIP3/NF-κB/SMAD pathway in patients with immune thrombocytopenia. Br J Haematol. 2018;180(3):395-411. [DOI] [PubMed] [Google Scholar]
- 156.Zhu X, Wang Y, Jiang Q, et al. All-trans retinoic acid protects mesenchymal stem cells from immune thrombocytopenia by regulating the complement-interleukin-1β loop. Haematologica. 2019;104(8):1661-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li H, Guan Y, Sun B, et al. Role of bone marrow-derived mesenchymal stem cell defects in CD8( + ) CD28(-) suppressor T-lymphocyte induction in patients with immune thrombocytopenia and associated mechanisms. Br J Haematol. 2020;191(5):852-862. [DOI] [PubMed] [Google Scholar]
- 158.Gong X, Sun D, Li Z, Shi Q, Li D, Ju X. Three-Dimensional culture of umbilical cord mesenchymal stem cells effectively promotes platelet recovery in immune thrombocytopenia. Biol Pharm Bull. 2020;43(7):1052-1060. [DOI] [PubMed] [Google Scholar]
- 159.Zhang P, Zhang G, Liu X, Liu H, Yang P, Ma L. Mesenchymal stem cells improve platelet counts in mice with immune thrombocytopenia. J Cell Biochem. 2019;120:11274–11283. [DOI] [PubMed] [Google Scholar]
- 160.Wang X, Yin X, Sun W, et al. Intravenous infusion umbilical cord-derived mesenchymal stem cell in primary immune thrombocytopenia: a two-year follow-up. Exp Ther Med. 2017;13(5):2255-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ma L, Zhou Z, Zhang D, et al. Immunosuppressive function of mesenchymal stem cells from human umbilical cord matrix in immune thrombocytopenia patients. Thromb Haemost. 2012;107(5):937-950. [DOI] [PubMed] [Google Scholar]
- 162.Eleuteri S, Fierabracci A. Insights into the secretome of mesenchymal stem cells and its potential applications. Int J Mol Sci. 2019;20(18):4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang JM, Zhu XL, Xue J, et al. Integrated mRNA and miRNA profiling revealed deregulation of cellular stress response in bone marrow mesenchymal stem cells derived from patients with immune thrombocytopenia. Funct Integr Genomics. 2018;18(3):287-299. [DOI] [PubMed] [Google Scholar]
- 164.Houwerzijl EJ. Ultrastructural study shows morphologic features of apoptosis and para-apoptosis in megakaryocytes from patients with idiopathic thrombocytopenic purpura. Blood. 2004;103(2):500-506. [DOI] [PubMed] [Google Scholar]
- 165.Sun RJ, Shan NN. Megakaryocytic dysfunction in immune thrombocytopenia is linked to autophagy. Cancer Cell Int. 2019;19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weiss CN, Ito K. microRNA-22 promotes megakaryocyte differentiation through repression of its target, GFI1. Blood Adv. 2019;3(1):33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pedersen OB, Grove EL, Kristensen SD, Nissen PH, Hvas AM. MicroRNA as biomarkers for platelet function and maturity in patients with cardiovascular disease. Thromb Haemost. 2022;122:181–195. [DOI] [PubMed] [Google Scholar]
- 168.Deng L, Wang X, Jiang L, et al. Modulation of miR-185-5p expression by EBV-miR-BART6 contributes to developmental differences in ABCG4 gene expression in human megakaryocytes. Int J Biochem Cell Biol. 2016;81(Pt A):105-111. [DOI] [PubMed] [Google Scholar]
- 169.Iizasa H, Wulff BE, Alla NR, et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285(43):33358-33370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tang J, Liu ZY, Tang Y, Wang Y. Effects of Dicer1 targeted by EBV-miR-BART6-5p on biological properties and radiosensitivity of nasopharyngeal carcinoma. Hum Exp Toxicol. 2021;40(6):977-993. [DOI] [PubMed] [Google Scholar]
- 171.Wang D, Zeng Z, Zhang S, et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell proliferation through the LOC553103-STMN1 axis. Faseb J. 2020;34(6):8012-8027. [DOI] [PubMed] [Google Scholar]
- 172.Lu Y, Qin Z, Wang J, et al. Epstein-Barr Virus miR-BART6-3p inhibits the RIG-I pathway. J Innate Immun. 2017;9(6):574-586. [DOI] [PubMed] [Google Scholar]
- 173.Rowley JW, Chappaz S, Corduan A, et al. Dicer1-mediated miRNA processing shapes the mRNA profile and function of murine platelets. Blood. 2016;127(14):1743-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16(9):961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bray PF, McKenzie SE, Edelstein LC, et al. The complex transcriptional landscape of the anucleate human platelet. Bmc Genomics. 2013;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Clancy L, Beaulieu LM, Tanriverdi K, Freedman JE. The role of RNA uptake in platelet heterogeneity. Thromb Haemost. 2017;117(5):948-961. [DOI] [PubMed] [Google Scholar]
- 177.Laffont B, Corduan A, Plé H, et al. Activated platelets can deliver mRNA regulatory Ago2•microRNA complexes to endothelial cells via microparticles. Blood. 2013;122(2):253-261. [DOI] [PubMed] [Google Scholar]
- 178.Leierseder S, Petzold T, Zhang L, Loyer X, Massberg S, Engelhardt S. MiR-223 is dispensable for platelet production and function in mice. Thromb Haemost. 2013;110(6):1207-1214. [DOI] [PubMed] [Google Scholar]
- 179.Ulger Z, Aksu S, Aksoy DY, Koksal D, Haznedaroglu IC, Kirazli S. The adhesion molecules of L-selectin and ICAM-1 in thrombocytosis and thrombocytopenia. Platelets. 2010;21(1):49-52. [DOI] [PubMed] [Google Scholar]
- 180.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125-132. [DOI] [PubMed] [Google Scholar]
- 181.Lyu J, Zhao L, Wang F, et al. Discovery and validation of Serum MicroRNAs as early diagnostic biomarkers for prostate cancer in Chinese population. Biomed Res Int. 2019;2019:9306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen X, Hu Z, Wang W, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130(7):1620-1628. [DOI] [PubMed] [Google Scholar]
- 183.Sui T, Ma L, Li X, et al. [Plasma microRNA profile in immune thrombocytopenia: screening and verification]. Zhonghua Yi Xue Za Zhi. 2014;94(14):1083-1086. [PubMed] [Google Scholar]
- 184.Zuo B, Zhai J, You L, et al. Plasma microRNAs characterising patients with immune thrombocytopenic purpura. Thromb Haemost. 2017;117(7):1420-1431. [DOI] [PubMed] [Google Scholar]
- 185.Bay A, Coskun E, Oztuzcu S, Ergun S, Yilmaz F, Aktekin E. Plasma microRNA profiling of pediatric patients with immune thrombocytopenic purpura. Blood Coagul Fibrinolysis. 2014;25(4):379-383. [DOI] [PubMed] [Google Scholar]
- 186.Sun Y, Hou Y, Meng G, et al. Proteomic analysis and microRNA expression profiling of plasma-derived exosomes in primary immune thrombocytopenia. Br J Haematol. 2021;194(6):1045-1052. [DOI] [PubMed] [Google Scholar]
- 187.Garabet L, Ghanima W, Rangberg A, et al. Circulating microRNAs in patients with immune thrombocytopenia before and after treatment with thrombopoietin-receptor agonists. Platelets. 2020;31(2):198-205. [DOI] [PubMed] [Google Scholar]
- 188.Neunert C, Terrell DR, Arnold DM, et al. American Society of hematology 2019 guidelines for immune thrombocytopenia. Blood Adv. 2019;3(23):3829-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Provan D, Arnold DM, Bussel JB, et al. Updated international consensus report on the investigation and management of primary immune thrombocytopenia. Blood Advances. 2019;3(22):3780-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bolton-Maggs P, George JN. Immune thrombocytopenia treatment. N Engl J Med. 2021;385(10):948-950. [DOI] [PubMed] [Google Scholar]
- 191.Hernández-Sánchez JM, Bastida JM, Alonso-López D, et al. Transcriptomic analysis of patients with immune thrombocytopenia treated with eltrombopag. Platelets. 2020;31(8):993-1000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.