Abstract
The spike protein comprises one of the main structural components of SARS-CoV-2 because it is directly involved in the infection process and viral transmission, and also because of its immunogenic properties, as an inducer of the protective antibodies production and as a vaccine component. The occurrence of mutations in this region or in other the virus genome regions, comprises a natural phenomenon in its evolution. However, they also occur due to the selective immune pressure, to which the agent is continuously subjected, especially in the spike protein immunodominant regions, such as the RBD. Mutations in the spike protein can change the virus’ fitness, increasing its affinity for target cells, its transmissibility and its virulence. In addition, these mutations can giving it the potential ability to evade the protective antibodies action obtained from convalescent sera or vaccine origin, as well as those used in therapy, which may favor the virus expansion and compromise the infection control.
Five mutations N501Y, E484K/Q, K417N/T, L452R and T478K, located in the spike protein RBD, have had a greater impact because they are associated with new attributes developed by the virus, which characterize the emerging variants of concern (VOCs) of SARS-Cov-2 identified so far. The occurrence of these mutations induces complex physicochemical effects that can alter the spike protein's structure and its function, which in turn, lead to changes in the agents' fitness. This manuscript discusses the attributes of VOCs associated with the physicochemical effects caused by the aforementioned mutations.
Keywords: SARS-CoV-2, Mutation, Receptor binding domain, Physicochemical effects, Variant of concern
1. Introduction
SARS-CoV-2 is a positive single-stranded RNA virus with a length of approximately 30,000 nucleotides, which encodes four structural proteins: spike protein [S], envelope protein [E], membrane protein (matrix) [M], and nucleocapsid protein (Kim et al., 2020). Each of them has specific functions in the viral cycle (Satarker and Nampoothiri, 2020).
Protein S is the fusion component that forms trimers on the virion surface. Composed of two subunits, S1 and S2, the S1 subunit is responsible for binding to the host cell receptor and the S2 for fusion with the cell membrane (Walls et al., 2020; Wrapp et al., 2020). The S1 subunit primarily contains the receptor binding domain (RBD), amino acid position 319–541, which interacts with host cell receptor angiotensin converting enzyme 2 (ACE2) (Satarker and Nampoothiri, 2020). This interaction triggers the protein S cleavage between the S1 and S2 fragments by cellular proteases found in the lung alveoli, such as transmembrane protease serine kinase 2 (TMPRSS2), thus initiating the virus entry into recipient cells and, consequently, the infection transmission to neighboring cells (Wan et al., 2020).
Protein S is also the main virus surface antigen as it has the property of inducing the host to produce neutralizing antibodies against SARS-CoV-2. Although most viral proteins are capable of inducing specific anti-SARS-CoV-2 antibodies, those targeting protein S are the only ones capable of blocking the virus entry into the host cell, interfering in the interaction of protein S with the receptor (ACE2) or interrupting the fusion mechanisms that the virus uses to access the host cell's cytoplasm (Lucas et al., 2021; Chi et al., 2020a; Brouwer et al., 2020). In addition, antibodies used in rehabilitation (based on plasma transfusion) and SARS-CoV-2 infected individuals therapy are also directed against protein S (Shen et al., 2020).
Finally, protein S is the viral component that serves as the basis for the development of all anti-SARS-CoV vaccines produced to date, offering as target sequences, full-length S protein, S1 and RBD, formulated with various adjuvants (Silveira et al., 2021; Shahzamani et al., 2021).
Due to these properties, protein S is of notable interest for being directly involved in the virus transmission mechanism, for being responsible for the antibodies induction that neutralize the virus and, finally, for its role as an immunogen, inducing protective antibodies against the virus. Therefore, the preservation of its primary structure is essential for maintaining its function. However, the SARS-CoV-2, as with viruses in general, is subject to evolution which can be characterized by the mutations emergence that can occur in any region of its genome (Singh et al., 2021). These mutations offer mechanisms for it to adapt to a new host and/or escape the immune response (cellular and humoral), or even become resistant to control measures against its propagation and to the therapeutic strategies adopted to contain the infection caused by it. The COVID-19 pandemic, characterized by the high transmission and replication rate of SARS-CoV-2, favored this agent to rapidly accumulate a constellation of mutations, giving it significant genetic diversity, since its initial detection in Wuhan, China, in December 2019 (Zhu et al., 2019). In particular, in the sequences encoding the spike protein. As the spike protein plays important properties in the virus biology, mutations that occur in it, especially in the RBD region, can compromise its structure and also its function and cause a great biological impact by altering the virus antigenic profile. Hence, it became critical to closely monitor the spike protein antigenic evolution in circulating viruses, especially in the pandemic context (Li et al., 2020).
As a product of this monitoring, many biologically significant mutations in protein S have been identified, and it appears that the RBD and N-terminal domain (NTD) located in this protein are mutational hotspots because they are under intense selective immune pressure (Greaney et al., 2021). Five mutations N501Y, E484K/Q, K417N/T, L452R and T478K, located in the spike protein RBD, have had a greater biological impact because they were associated with new attributes developed by the virus, which served as a basis for characterizing the variants of concern (VOC) of SARS-Cov-2 that emerged in the pandemic context. Other mutations such as P681H, D614G and the 69/70 deletion, located in regions outside the RBD and which also have implications for the virus fitness, have been identified. The occurrence of these mutations induces complex physicochemical effects that can alter the spike protein's structure and its function.
Several molecular dynamics studies have demonstrated these effects and their implications for the SARS-CoV-2 variants fitness (Alaofi and Shahid, 2021; Istifli et al., 2021; Peters et al., 2021). However, in most of these studies the change in the variants' fitness is demonstrated considering the combined mutations effect (or the set of them) identified in the VOCs, which limits the understanding of the effect and impact caused by each one of them individually. In this manuscript, it was proposed to associate the physicochemical effect of each mutations mentioned above with the agent's fitness, as well as the attributes presented by the VOCs.
1.1. Physicochemical and biological effects of the mutations N501Y, E484K/Q, K417N/T, L452R and T478K on spike protein
-
1.
N501Y mutation
Mutation first identified in the SARS-CoV-2 variant that emerged in the UK (lineage B.1.1.7) (see below) (Rambaut et al., 2020), and was subsequently identified in the variants that emerged in South Africa (B.1.351) and in Brazil (P.1).
The N501Y mutation is characterized by the substitution of an asparagine (N) residue with a tyrosine (Y) (N501Y) residue. It is located within RBD in the spike protein S1 region, specifically in a region called the Receptor Binding Motif (RBM), amino acid position 437–508 (Satarker and Nampoothiri, 2020). The residue at this position is one of the six major virus-cell contact residues present within that domain (Lim et al., 2020). The tyrosine residue introduction forms an extra hydrogen bond with the angiotensin converting enzyme 2 receptor (ACE2) (Lys353-Tyr501), which increases the surface protein's electrostatic docking attraction, and therefore, the SARS-CoV-2 virus binding affinity to the host cell (Khan et al., 2021). In other words, this mutation induces a lower virus-cell dissociation constant, as it causes a stronger binding between the new variant and the receptor when compared to the wild type, according to molecular dynamics studies (Tian et al., 2021).
Structural biological studies of the SARS-CoV-2 RBD also concluded that in addition to increasing the binding strength of the RBD to human ACE2 (Starr et al., 2020a), the N501Y mutation may also contribute to the spike protein maintenance in a so-called “open” prefusion conformation state, promoting greater efficiency in viral entry, contributing to greater the agent infectivity (Teruel et al., 2021). This prefusion isoform of the protein S is the necessary state for the RBD to recognize and engage the ACE2 receptor. The same effect has been predicted for other mutations on glycine residues (404, 416, 504) as well as at K417 residue (Teruel et al., 2021). Therefore, the most prominent biological effect of this mutation is to attribute to variant greater ability to transmit and propagate in the environment, which was proven based on its rapid expansion and the N501 lineage replacement (Leung et al., 2021). However, it was suggested that the N501Y mutation alone would not be sufficient to give the virus a different fitness from that observed in the wild-type virus (Khan et al., 2021). Additionally, it was also suggested that this mutation had no potential to compromise the vaccines efficacy already developed against the agent (Conti et al., 2021), although its presence reduces the neutralizing activity of anti-RBD antibodies from convalescent sera (Lu et al., 2021).
-
2.
K417N mutation
Characterized by the replacement of a lysine (K) residue with a arparagine (N) residue (K417N), this mutation, although it does not contribute to binding toACE2 receptor (since it is outside the RBM), occurs in an epitope for neutralizing antibodies (Tada et al., 2021a). Its is suspected that its emergence is associated with the potential for escape developed by VOCs from neutralization by various monoclonal antibodies classes and also from convalescent sera, which have been shown protection against severe forms of COVID-19 (Yuan et al., 2021). Additionally, others studies have shown that this mutation may also contribute to escape neutralization by antibodies elicited by vaccines Pfizer-BioNtech BNT162b2 and Moderna mRNA-1273 (Tada et al., 2021a).
It has been shown that the physicochemical effect caused by the K417N mutation is the binding strength reduction RBD-ACE2, as a consequence of the salt-bridge loss between K417 and D30 (located in ACE2) when K417 changes to N417 and forms a pair between K484 and E75 (also located in ACE2), increasing virus-cell dissociation constant (Jawad et al., 2021). This effect could minimize the virus fixation potential in the cell and, thus, decrease its infectivity. But, interestingly, the co-occurrence of the N501Y mutation (which is observed in 95% of cases (Yuan et al., 2021)), re-establishes the variant's capacity to efficiently interact with the host cell, restoring the potential for infection. Studies by Kim Y et al. (2021) have also shown that the K417N mutation, when in association with the E484K mutation, dramatically increases virus-cell fusion (compared to wild-type virus or the N501Y variant) as the E484K mutation improves the electrostatic complementarity with the negatively charged ACE2, through the change from a carboxylic acid (E) to an amine group (K). This physicochemical effect may confer greater transmissibility to SARS-CoV-2, although the infectivity remains similar (Kim et al., 2021). Therefore, despite the presence of the K417N mutation alone has a negative effect on the infection clinical course, the variant's attributes are reset by co-occurrence of other mutations.
Due to its location, it has been suggested that the K417N mutation may have been selected as a mechanism for evading the humoral response (Wang et al., 2021a), which justifies the ability to escape from neutralizing antibodies action by variants that express this mutation, such as those that emerged in South Africa (B.1.351) and in Brazil (P.1).
A second mutation was identified at this position, characterized by the substitution of the lysine residue with a threonine (K417T). It has also been found in association with the N501Y and E484K mutations. Despite being different from each other, both give similar attributes to the viral variants, although variations in the binding pattern are noted due to the different biochemical characteristics that exist among the mutated residues (Khan et al., 2021).
-
3.
E484K mutation
The E484K mutation comprises the substitution of a residue with a glutamate (E) with a lysine (K) at position 484 (E484K) in the RBD. The amino acid at that position serves as a contact point for ACE2 (Lan et al., 2020). This exchange results in the introduction of an additional hydrogen bond between the spike glycoprotein and the SARS-CoV-2 RBD (Glu35-Lys484). This extra bridge decreases the ACE2-RBD dissociation constant by increasing the binding strength of the ACE2-E484K mutant complex, giving the variant greater cell surface docking capacity than that observed in wild-type virus (Khan et al., 2021). Besides that, the E484K mutation also causes the conformational rearrangements of the loop region containing the mutant residue, which leads to tighter binding interface of RBD with ACE2 and formation of some new hydrogen bonds. The tighter binding interface and the new hydrogen bonds formation also contribute to the improved binding affinity of RBD to the receptor ACE2 (Wang et al., 2021b). This substitution may be responsible for the rapid spread and high infectivity of the SARS-CoV-2 variant that presents it, especially when associated with the N501Y mutation (Zhao et al., 2021). In addition, since position 484 is part of the most important site for virus recognition by neutralizing antibodies, the E484K mutation occurrence also significantly reduces the binding affinities to RBD for some neutralizing antibodies/nanobodies, mainly owing to the unfavorable electrostatic interactions caused by it (Wang et al., 2021b).
Therefore, the occurrence of mutation in this position is a matter of great concern to public health since it favors the virus spread, compromises the neutralizing activity of therapeutic monoclonal antibodies (Weisblum et al., 2020) and also gives the virus a ability to evade immunity induced by natural infection and by vaccination (Wang et al., 2021b; Jangra et al., 2021; Liu et al., 2021a). Accordingly, it has been suggested that mutations in this position can reduce by up to 10 times the virus recognition capacity by therapeutic monoclonal antibodies and also polyclonal ones (originated from natural exposure to SARS-CoV-2) (Greaney et al., 2021). Additionally, it is important to note that this position along with other positions with interface residues are the main access points for drug discovery against SARS-CoV-2 variants (Yang et al., 2021), which means that mutations on that site could compromise its effectiveness. The E484K mutation is present in the South African, Brazilian and emerging Indian variants, B.1.351, P.1 and B.1.617 lineages, respectively.
Another mutation at the same position (E484Q) has been reported in some variants, although its effects are still poorly understood. One of them appears to be the disruption of an electrostatic binding in the residue E484 of the spike protein RBD with K31 at the ACE2 interaction interface. Structural analysis of the effect of this mutation revealed a decrease in intramolecular and intermolecular contacts in this region, compared to wild type, with sufficient potential to destabilize the RBD-ACE2 binding (Cherian et al., 2021). However, the L452R residue mutation (hydrophobic L452 to the hydrophilic 452R) which often occurs concurrently with E484Q, can help in interactions with water molecules and in the general re-stabilization of the complex, maintaining high binding avidity (Cherian et al., 2021; Augusto et al., 2021a).
Finally, it has been shown that E484Q can decrease the binding capacity of some monoclonal antibodies to variant strains, compared to the wild-type strain (Cherian et al., 2021), as well as of antibodies induced by natural infection or vaccination, as evidenced by inhibition assays (Augusto et al., 2021a). Despite the concurrence of this mutation with the L452R (discussed bellow), there seems to be no synergism in the effect of these two mutations in relation to the neutralizing capacity of the variant by anti-RBD antibodies (Ferreira et al., 2021).
-
4.
L452R mutation
It has been suggested that the L452R mutation emergence is a consequence of the SARS-CoV-2 adaptive evolution, in response to either the epidemiological containment measures or due to a growing proportion of the population with immunity to the original viral variants, i.e. the reconvalescents and vaccinated individuals (Tchesnokova et al., 2021). Although the L452R functional impact has not yet been elucidated, it is known that leucine-452 is positioned in the RBM (within the RBD), in the direct contact interface with the ACE2 receptor, despite this residue does not directly contact the receptor (Lan et al., 2020).
It has been shown that the substitution of leucine (L) with a arginine (R), in association with the E484Q mutation results in an increase in the binding energy between the RBD-ACE2 complex, predicting stronger virus-cell binding (Kumar et al., 2021). This physicochemical effect would facilitate the SARS-CoV-2 penetration into the cell, giving it greater infectivity and transmissibility (Chen et al., 2020a). Furthermore, it was observed that its occurrence together with the T478K mutation increase the stability and intra-chain interactions in the spike protein, which may change the interaction ability of neutralizing antibodies (Kumar et al., 2021).
In order to prove the individual effect of this mutation on virus fitness, pseudoviruses were generated carrying the L452R mutation with D614G and with the D614G mutation only. It was observed that the pseudoviruses generated carrying the L452R mutation demonstrated greater ability to penetrate cells that express the ACE2 receptor and the TMPRSS2 cofactor for SARS-CoV-2 when compared to the pseudoviruses that had only the D614G mutation. This finding proposed the association of the L452R mutation with the increased infectious capacity of the variant in which the mutation is detected (Deng et al., 2021).
The individual influence of the L452R mutation was also proven in the immunity context. Studies have shown that this mutation gives the variant moderate resistance to neutralization by antibodies elicited by previous infection or vaccination (Deng et al., 2021; Mor et al., 2021), which obviously gives L452R mutants potential to also evade humoral immunity. Furthermore, it has been shown that shifts at position 452 can also influence the cellular immune response against SARS-CoV-2. The region that spans 448–456 of the S protein comprises the epitope presented by HLA-A24 (Kiyotani et al., 2020), which comprises the region of recognition by CD8+ lymphocytes (Motozono et al., 2021). Accordingly, it has been suggested that the L452R mutation has potential to confer resistance to antigen recognition by HLA-restricted cellular immunity, compromising the CD8+ cells activation. This property enables this variant to escape HLA-restricted cellular immunity (Motozono et al., 2021), leading to the loss of immune control, which would favor the infection progression (Le Bert et al., 2020). Finally, it has also been shown that the L452R mutation significantly improves viral replication capacity by increasing binding affinity to human ACE2 as well as S protein stability (Motozono et al., 2021).
-
5.
T478K mutation
The T478K mutation is also located at the spike/ACE2 interaction interface. Its expansion is worryingly occurring among SARS-CoV-2 sequences collected since early 2021. It has been prevalently identified in circulating strains in Mexico, but also in North America and European countries such as Germany, Switzerland and Sweden (Di Giacomo et al., 2021).
Characterized by the replacement of threonine with a lysine, this mutation increases the spike protein electrostatic potential by inserting a basic residue, lysine (K), in place of a polar but neutral residue, threonine (T), making the protein surface more positive, which could influence the protein interaction with the cellular receptor (Di Giacomo et al., 2021). This interaction capacity has been evaluated by binding free energy (BFE) of S protein and ACE2, generated by mutations. Different mutations can generate different BFE, which can range from 0.01 kcal/mol (or less) to more than 0.999 kcal/mol. The higher the BFE, the greater the binding strength (Wang et al., 2022). It has been shown that the T478K mutation promotes a change in BFE of S protein and ACE2 near to 1.00 kcal/mol, so very high, which strongly enhances the binding of the RBD–ACE2 complex (Cherian et al., 2021; Wang et al., 2022), and thus the infectivity of the variant. However, in a deep mutational scanning of the SARS-CoV-2 RBD, the T478K mutation did not have a significant effect on binding to human ACE2 (Starr et al., 2020a; Rodríguez-Maldonado et al., 2021). Despite conflicting data, the emergence of this mutation has made the variant more infectious (Wang et al., 2022). In addition, the T478K mutation may be involved in immune evasion, particularly escape from antibody neutralization (Rodríguez-Maldonado et al., 2021), or in the interaction impairment of RBD with drugs (Di Giacomo et al., 2021).
1.2. Other relevant mutations in the spike protein located outside the RBD region
-
1.
Deletion 69-70
Mutation identified so far only in the SARS-CoV-2 variant that emerged in the UK (lineage B.1.1.7). Consequence of deletion of 21,765–21770 genome region, it is characterized by the two amino acids deletion at positions 69/70 (del69-70/(ΔH69/V70), and appears to have arisen in multiple genetic backgrounds of SARS-CoV-2, accordingly with some studies (Ibba et al., 2021; Borges et al., 2021).
Amino acids 69 and 70 are located in the NTD of the spike protein S1 subunit which has also been associated with the neutralizing antibodies induction against SARS-CoV-2 (Chi et al., 2020b). It has been suggested that deletion of these residues may allosterically alter the S1 region conformation (Wrapp et al., 2020), and induce non-recognition of this region by certain classes of specific NTD monoclonal antibodies (Diamond et al., 2021).
Its occurrence also has a great impact on the infection laboratory diagnosis, leading to what has been termed “spike gene target failure” (SGTF) or “spike gene drop out” (Borges et al., 2021). This denomination is associated with the fact that this mutation may cause failure in the gene S sequences detection, or cause late amplification of its targets by some PCR assays that amplify this region, producing a negative result (Borges et al., 2021; Public Health England, 2021a).
-
2.
P681H mutation
The P681H mutation was first identified in the UK variant (20I/501Y.V1) (Rambaut et al., 2020). It is not located in the NDT or RBD domains of the spike protein S1 region, but rather immediately adjacent to the proteolytic cleavage site for furin and furin-like proteases, between the arginine and serine residues, specifically at the junction of the binding (S1) and fusion (S2) domains to the surface protein receptor (Polg á r, 1989). The S1/S2 cleavage comprises a phenomenon subsequent to the virus spike interaction with the host cell membrane, and it needs to occur to promote the virus entry into the respiratory epithelial cells, being essential for sustained transmission of SARS-CoV-2, according to experiments in animal models (Peacock Thomas et al., 2020; Zhu et al., 2020).
It has been suggested that the P681H mutation occurrence may increase protein S cleavage by furin-like proteases, which predicted increase membrane fusion (Yarmarkovich et al., 2020) and enhance systemic infection (Huang et al., 2020). In order to assess whether the effect of this mutation could facilitate the virus penetration into the cell, potentiating the infection, functional assays in cell culture were carried out. Using like-virus particles harboring SARS CoV-2 spike proteins, it was shown that between variants expressing the P681H mutation and those that do not, there did not appear to be significant differences in cell penetration capacity, despite differences in cleavage potentials. This finding suggested that this mutational phenomenon does not significantly affect viral entry or its cell-to-cell spread (Lubinski et al., 2021), except in cases with low expression of the ACE-2 receptor, as suggested by Dicken et al. (2021) (Dicken et al., 2021). Since the results obtained could not fully confirm the relationship of this mutation with the attribute of the variant, it was deduced that this mutation would not be the only one responsible for the increase in the spread potential of the variant (Lubinski et al., 2021), which led to believe that other factors could explain the increase in the variant transmission rates that presented this mutation.
More recently, another mutation at the same position, identified as P681R, has emerged in the Delta variant (see below), identified in India. It was demonstrated that this substitution clearly increased spike processing, a phenomenon that likely conferred an advantage in replication fitness observed in the Delta variant. Accordingly, laboratory assays have shown that after infection of respiratory epithelial cells by this variant, the P681R mutation would facilitate cleavage into S2 by the cell surface protease (TMPRSS2), leading to the S2 virus-plasma membrane fusion peptide activation (Saito et al., 2021), producing, however, only an effect similar to that of the P681H mutation. Additionally, an association of the P681R mutation with increased transduction of the variant (in Vero-TMPRSS2 cells) (Lubinski et al., 2021) and improvement in viral replication compared to the original virus, was also observed (Liu et al., 2021b), suggesting an increase in infectivity (Lubinski et al., 2021). However, there is evidence that this substitution alone would not be sufficient to promote all these effects, suggesting that its functional consequences depend on its emergence in the other spike protein mutations context (Lubinski et al., 2021).
-
3.
D614G mutation
This mutation was rare globally. However, it gained prominence when the virus spread from Asia to Europe and USA, quickly becoming the dominant form in the world (Yurkovetskiy et al., 2020). It is present in all the variants considered as “variant of concern” (VOC), characterized below and identified so far, grouping them within the same clade. This suggests that this mutation emerged as result of a natural selection process experienced by the agent (Giovanetti et al., 2021).
The D614G mutation is located on the spike protein protomer surface, where the formation of a hydrogen bond between protomers occurs, joining a residue from the S1 unit of one protomer to the residue from the S2 unit of the other protomer. It is characterized by the exchange of an aspartate (D) residue with a glycine (G) as a consequence of the A to G mutation at nucleotide 23,403 of the virus genome, with reference to the Wuhan viral strain (Korber et al., 2020).
The G614 mutation has implications for the spike protein structure, as it eliminates hydrogen bonds between the protein's S1 unit and S2 units naturally present in D614. This physicochemical change increased the protein chain flexibility (Korber et al., 2020), confering enhanced thermodynamic stability irrespective of conformational states and most optimal for binding host ACE-2 (Yazhini et al., 2021). Therefore, it is assumed that this mutation has improved the transmission efficiency of SARS-CoV-2 (Yazhini et al., 2021; Volz et al., 2021) by up to approximately 50%, according to Pearson et al. (2021) (Pearson et al., 2021), and contributed to the increase in the variants' infectivity (Giovanetti et al., 2021). Additionally, it has been suggested that this mutation would be associated with an increase in the fatal cases rate (Becerra-Flores and Cardozo, 2020), a phenomenon clearly observed after the VOCs emerging, so far.
Another important biological effect caused by this mutation is the low cycle threshold detected in RT-PCR assays, suggesting higher viral load in the infected patients upper respiratory tract, and, therefore, association of the G614 mutation with a more infectious virus (Groves et al., 2021). On the other hand, this mutation does not appear to be the cause of greater resistance to neutralization by convalescent sera or by vaccination-induced antibodies, since it is not located in the spike protein RBD, but in the interface between the individual spike protomers that stabilize the protein trimeric form on the virion surface (McAuley et al., 2020).
In summary, the emergence of this mutation led to an increase in viral fitness for replication in the respiratory tract, improved binding to the ACE2 receptor and conformational changes within the spike protein (Gobeil et al., 2021; Ozono et al., 2021), which would justify the rapid global expansion of SARS-CoV-2 variants.The Fig. 1 presents a summary of the mutations addressed in this manuscript, the physicochemical effect caused by it on the protein S structure, the implication of its occurrence in the virus' fitness and, finally, the attribute aggregated by the variant due to its emergence.
Fig. 1.
Physicochemical effect of the mutations in RBD and adjacent regions of protein S and biological effect/attributes of the VOCs.
1.3. Characterization of the variants of concern
Variants of Concern (VOCs) comprise emergent variants of SARS-CoV-2 that harbor mutations in the genome that, depending on its location, can impact in the virus transmission, its virulence and in the vaccinal or naturally developed immunity, compromising the control measures adopted (social and public health) to contain the agent (CDC, 2021a).
Since the fall of 2020, 5 VOCs have emerged globally, and are the current focus of epidemiological, clinical and virological investigation. They are commonly referred to by the country in which they were originally identified. They are: the 20I/501Y.V1 variant (lineage B.1.1.7), which appeared in the United Kingdom; 20H/501Y.V2 variant (lineage B.1.351), emerged in South Africa; 20J/501Y.V3 variant (Lineage P.1, originating from B.1.1.28), emerged in Brazil/Japan; G/478K.V1 variant (Lineage B.1.617.2), emerged in India (CDC, 2021a), and a novel variant recently classified, B.1.1.529, which also emerged in South Africa (CDC, 2021a). Several other variants are under investigation (Public Health England, 2021a, 2021b). The first four VOCs have also been designated as Alpha, Beta, Gamma and Delta, respectively, by the WHO as of May 31, 2021 (World Health Organization, 2021a), and the fifth VOC was designated as Omicron, on November 26, 2021 (World Health Organization, 2021b). In each of these variants, the spike protein contains mutations clustered in its RBD and in the NTD region.
-
1.
20I/501Y.V1 variant
Also known as VOC202012/01 (Lineage B.1.1.7). It emerged in the United Kingdom, being identified in September 2020 (Rambaut et al., 2020). Its emergence was directly associated with the SARS-CoV-2 infections resurgence in South London, where it quickly became the predominant strain (Galloway et al., 2021). Since then, this variant has been detected in several countries around the world, including the USA.
It has an unusually large number of mutations in the genome (17 mutations) among which are included three deletions, 7 missense mutations, in addition to three impact mutations, N501Y and D614G and the 69–70 deletion (Rambaut et al., 2020; Starr et al., 2020b), whose effects on virus properties are described above. Evidence has been presented that this variant was the cause of increased hospitalization and risk of death in the UK compared to other variants (Challen et al., 2021). Its emergence was also associated with increased virus transmissibility (Challen et al., 2021).
The 20I/501Y.V1 variant does not appear to have had a high impact on the some vaccines efficacy. There was also evidence indicating that sera from patients immunized with the Pfizer-BioNTech SARS-CoV-2 or modern vaccine (m-RNA-1273) maintain the ability to neutralize it, in vitro (Muik et al., 2021; Wu et al., 2021). In addition, other studies have shown that this variant is vulnerable to neutralization by convalescent serum and by antibodies generated with nanoparticle vaccine (NVXCoV2373, Novavax) (Wang et al., 2021c; Shen et al., 2021). Furthermore some therapeutic monoclonal antibodies retain the ability to neutralize this variant (Tada et al., 2021b).
-
2.
20H/501Y.V2 variant
Variant from the B.1.351 lineage that emerged in South Africa, and was subsequently isolated in neighboring countries as well. Despite having emerged independently from B.1.1.7, it shares some mutations with that variant, including E484K (present in some strains) and N50IY, in addition to the D614G mutation. Other mutations identified in the spike protein are L18F, D80A, D215G, L242-244del, R246I and K417N. There are no 69/70 deletions (CDC, 2021a). It has been highlighted that this variant, as well as the Brazilian one (described below), is more lethal than the UK one due to the three mutations combination mentioned above (K417N, E484K, N501Y), rather than a single one (N501Y), demonstrated by the 20I/501Y.V1 variant (Khan et al., 2021). The E484K substitution is characteristic of the B.1.351 strain, but is also present in P.1, P.2 (B.1.1.28 strain variants). It arose independently in many genomic contexts other than SARS-CoV-2, including some strains of the B.1.1.7 lineage (Moustafa et al., 2021).
This variant was responsible for the 'second wave' in South Africa. Its emergence has been associated with increased transmissibility, immune escape, or a combination of the two attributes (Pearson et al., 2021). It has also been shown to be refractory to neutralization by most anti-NTD monoclonal antibodies (mAbs), and also by several mAbs targeting the binding receptor on the RBD. Furthermore, it has been shown to be more resistant to neutralization by convalescent plasma (∼11–33 times) and vaccinated sera (∼6.5–8.6 times) (Ho et al., 2021). Similar effects were observed from other studies, with the B.1 variant as a reference (Edara et al., 2021). This effect, in large part, has been attributed to the E484K mutation.
Finally, one study showed reduced plasma neutralizing activity against SARS-CoV-2 variants encoding E484K, N501Y, or K417N:E484K:N501Y (characteristic combination of this VOC) in a cohort of 20 volunteers who received either the Moderna (n = 12) or Pfizer (n = 6) vaccine, 8 weeks after the second dose (Wang et al., 2021a). Similar results were seen with sera from Moderna-vaccinated individuals (Edara et al., 2021).
Although studies have shown isolated effects of each of the three mutations, it has been suggested that the attributes developed by this variant result from the combination of these mutations and not just a single one (Cele et al., 2021; Wibmer et al., 2021).
-
3.
20J/501Y.V3 variant
Also known as P.1 (a branch of the B.1.1.28 lineage), phylogenetic analysis indicated that P.1 and another lineage, P.2 (Candido et al., 2020) were descendants of lineage B.1.1.28 that was first detected in Brazil in early March 2020. Concurrently, cases of SARS-CoV-2 P.1 infection were reported in Japan in travelers from Amazonas (Fujino et al., 2021). It contained 17 amino acid changes (including 10 in the spike protein - L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, and T1027I, compared with its immediate ancestor (B.1.1.28), three deletions, four synonymous mutations, and a four base-pair nucleotide insertion compared with the most closely related available sequence (GISAID ID: EPI_ISL_722,052) (Faria et al., 2021).
Within the region in which it propagated, it was estimated that this variant could be between 1.7 and 2.4 times more transmissible than local non-P.1 strains and that it could have a potential varying between 21 and 46% to evade protective immunity elicited by previous infection with non-P.1 strains (Faria et al., 2021). Accordingly, there is evidence that the ability of antibodies generated by natural exposure (Gidari et al., 2021) or vaccination (Anderson et al., 2020) to recognize and neutralize the virus may be affected by aggregated mutations in this variant. In addition, it was also demonstrated that the neutralizing activity of monoclonal antibodies directed against the RBD and NTD region of the P.1 variant were markedly or completely abolished, including of two potent mAbs targeting the receptor-binding motif (Wang et al., 2021d). The results in P.1 mimic those observed for B.1.351, which should not be surprising since the triple RBD mutations in P.1 and B.1.351 are basically the same.
-
4.
G/478K.V1 variant
The variant of interest B.1.617 emerged in India and comprises three distinct sublineages (B.1.617.1, B.1.617.2, and B.1.617.3) with different mutational profiles. Among them, only sublineage B.1.617.2, or Delta, was confirmed as a VOC by the WHO (World Health Organization, 2021c) and is now internationally recognized as such for the clinical-epidemiological impact it has caused since its emergence (Lazarevic et al., 2021). This variant has been associated with the second wave of COVID-19 in India (Vaidyanathan, 2021). Its is characterized by 13 amino acid changes, contain diverse mutations in the NTD and the RBD of the SARS-CoV-2 spike protein, but four in its spike protein, which are currently of particular concern. They are: L452R, T478K, E484Q, and P681R (Shiehzadegan et al., 2021).
To this variant has been attributed a greater potential for transmissibility when compared to other pre-existing VOCs circulating in Europe and other countries (Davies et al., 2021) and data from GISAID (https://www.gisaid.org/) has confirmed this claim. According to Public Health England, 2021 (PHE), it is at least 40–50% more transmissible than the B.1.1.7 variant, and it is on the rise worldwide (Kirola, 2021).
It is suspected that the greater transmissibility potential of the B.1.617 variant is associated with multiple substitutions of neutral or negatively charged amino acids for positively charged amino acids in the protein S structure (Pascarella et al., 2021). But this peculiarity has long been directly associated with both the E484Q and L452R amino acid substitutions (Chen et al., 2020b), cited above. Despite this fact, the Delta variant is the most infectious variant among all variants formally named by the WHO due to the combined effect of L452R and T478K (Chen et al., 2021).
Other important mutations found in the variants described above have also been identified in this variant, such as N501Y, K417N and D614G (Baral et al., 2021), whose effects on virus fitness have already been discussed. The association of these mutations has given the Indian variant a greater potential for escape (partial or total) from neutralization by some monoclonal antibodies, which makes its resistance to therapeutic intervention possible (Planas et al., 2021). Furthermore, the neutralization of this variant by plasma panels collected from convalescent individuals was reduced when compared to the neutralizing capacity of the original virus. These findings demonstrate the inability of these antibodies to block the interaction between ACE2 and the RBD of this variant (Augusto et al., 2021b). Finally, it has been shown that there are significant reductions in neutralizing titers of sera collected from Oxford-Astra Zeneca and Pfizer-BioNTech vaccine recipients (Planas et al., 2021; Liu et al., 2021c).
-
5.
B.1.1.529 variant
Variant classified by WHO as VOC on November 26, 2021, which also emerged in South Africa. The mutation profile includes 32 spike mutations, including in the RBD and furin cleavage site, and additional mutations outside spike of uncertain significance (Public Health England, 2021b). The genome also contains the spike deletion at the position 69–70, up to now found only in B.1.1.7 variant, in addition to mutations K417N, T478K, N501Y, D614G, all already identified in the previous variants, and also a mutation at the residue position 484 (A484), the effect of which is still uncertain. The SGTF, which is a consequence of del69/70, has been used as a marker to define “highly probable case”, “probable case” and “possible case” of infection by Omicron, in association with other criteria (Public Health England, 2021b). An N-gene drop out, also referred to as an N-gene target failure (NGTF), due to a nine-nucleotide deletion in the N-gene, spanning positions 28370–28362, has also been a finding associated with omicron variant infection, and its occurrence is used for virological characterization. This deletion can also compromise virus detection by some genomic amplification tests (CDC, 2021). From the genomes available so far, the shared mutation profile is: S: A67V, Δ69–70, T95I, G142D/Δ143-145, Δ211/L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F (Public Health England, 2021b).
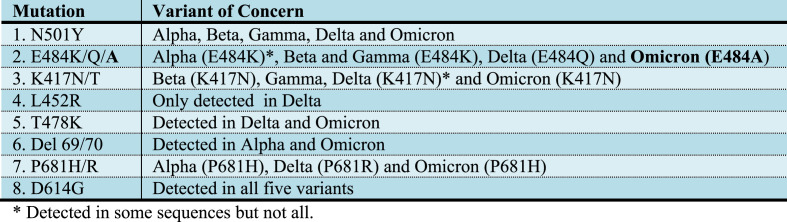
Based on location in the genome, structural modelling and experience from other variants, it was predicted that these mutations could change the behaviour of the virus with regards to immune escape, transmissibility, and susceptibility to some treatments, particularly therapeutic monoclonal antibodies (Public Health England, 2021b). Concordly, it has already been shown that some of them, specifically located in the RBD region, such as S477N, Q493R, G496S and Q498R, result in additional H-bonds and salt bridges, resulting in a more positive RBD surface, complementary to the negatively charged ACE2 interaction interface. This physicochemical effect increases the RBD-ACE2 interaction and supposedly the infectivity/transmissibility of the variant (Queirós-Reis et al., 2021). Interestingly, it has also been reported that other mutations, also exclusive to omicron, conferred attributes similar to those developed by other VOCs, in addition to inducing physicochemical effects similar to those caused by already known mutations (Shah and Woo, 2022). As expected, this variant has spread rapidly around the world since its emergence in Africa, demonstrating its potential for dissemination and infectivity. Fig. 2 presents a summary of the mutations discussed here and the variants in which they were detected.
Fig. 2.
Summary of the mutations and the variants in which they were detected.
2. Conclusion
The physicochemical effect caused by mutations at positions 501, 484, 417, 452 and 478 located in the RBD, but also in other regions of the VOCs spike protein, often seems to justify the change caused in the agent fitness and the attribute developed by the variant.
The increase in the spike protein's electrostatic potential, caused by the N501Y or T478K mutation, leads to greater affinity in the ACE2-RBD interaction, developed by the agent. The conformational change in protein S, as a consequence of the E484K, L452R mutation or the 69/70 deletion, justifies the non-recognition of the variant by specific antibodies. Or, the increase in fusion potential provided by the K417N/E484K or L452R mutations, or the increase in cleavage potential provided by P681H may justify greater variant-cell affinity. The change in the agent's fitness gives the variant new attributes characterized by an increase in its transmission capacity and viral load in the infection; ability to evade the neutralizing antibodies action arising from natural exposure or vaccination; non-response to treatment with monoclonal antibodies or convalescent serum, immune system evasion and compromised diagnosis. All these alterations and new attributes converge in order to increase the agent's virulence, provide greater infection severity, increase hospitalizations cases and induce greater the infection lethality.
It is remarkable that despite the different positions in which mutations occur and the differences between them, the physicochemical effect observed is similar between some of them. This observation suggests that the change observed on the virus’ fitness and the specific attribute (or a set of them) developed by a variant seem to be the consequence not of a specific mutation, but of a set of emerging mutations capable of inducing to the same physicochemical effect, and part of the studies referenced here demonstrate this fact.
This would partly explain why several attributes are common among different VOCs while having distinct impact mutations, some of them almost unique, although there are several mutations in common among the VOCs. However, it must be considered that the variants' attribute may also be influenced by the synergistic effect of specific mutations since they emerge in a constellation of other mutations whose impact is still unknown. Furthermore, the mutations discussed here may not be solely responsible for the variants' attributes. Finally, it should be considered that other mechanisms (virological, immunological or molecular) may also be involved in changing of the variants’ fitness, and the attribute developed is not only a consequence of the mutation physicochemical effect. Despite these possibilities, the isolated effect of each of the mutations on the fitness of SARS-CoV-2 variants needs further studies in order to identify its true impact, with deserved attention to the similar physicochemical effect caused by residues that have totally different physicochemical properties, as shown here.
In parallel with the emerging impact mutations addressed here, the evolutionary character of SARS-CoV-2 and its rapid adaptation to the environment, draw attention. After the first wave of COVID-19, the first VOC recognized as 20I/501Y.V1 (UK), had as its main attribute the increase in transmission rates, which was associated with the N501Y mutation, the most prominent in this variant, the despite other existing mutations. This variant showed little impact on therapeutic or vaccination-induced neutralizing antibodies action, since only a few sequences of this variant had the E484K mutation. The emerging in South Africa, even evolving independently of its predecessor, emerged conserving the N501Y mutation and presenting additional mutations E484K, K417N, which would confer new attributes to the mutant strain, including moderate impact on therapeutic antibodies and vaccine-induced or acquired by natural exposure, in addition to maintaining high transmission rates. Subsequently, P1, responsible for the second wave of COVID-19 in Brazil, emerged retaining the same mutational profile in the RBD presented by the emerging variant in South Africa, as well as the same attributes, in addition to being responsible for the high rates of hospitalizations and deaths. The fourth VOC, G/478K.V1, emerging in India, harbors some mutations different from those observed in the previous three, but has properties that contribute to the same biological effects, in addition to its ability to not only evade the action of specific neutralizing antibodies, but also, to confer resistance to the recognition of viral antigens by HLA-restricted cellular immunity, due to the L452R mutation emergence. And now, recently, the B.1.1.529 variant, also emerging in South Africa and Botswana, which demonstrates the majority of the mutations in the RBD region that emerged in previous variants, in addition to others whose physicochemical effect is still unknown, but keeping the same attributes of the previously identified variants.
The SARS-CoV-2 expansion through the emergence of new variants suggests that it is progressively evolving to the adapt to human species, in order to remain continuously viable in nature, despite the implemented control measures, including population vaccination. However, the evolutionary character based on constant mutations and emergence of new virulent variants, with exacerbated behavioral characteristics (transmissibility, infectivity, replication, evasion of neutralizing antibodies and induction of mortality), and also occurring in a short period of time, demonstrates an agent with little capacity of adaptation, which has brought the disease burden to the population whose impact could be greater without the vaccine. In this sense, at the moment there are no reasons to believe that changes of the virus have occurred in the direction of decreased pathogenicity to establish the expected balance with the human species and, at the same time its perpetuation in the nature, without causing great harm to the human population.This suggests that the COVID-19 pandemic may still stress our healthcare systems in unprecedented ways. If today the decrease in the infections severity is noticed, it has been related to the containment measures still maintained, capable of influencing the viral load magnitude that is transmitted, and which, associated with immune mechanisms triggered by natural or vaccine immunization, minimize the impact caused by the persistent virus circulation.
Funding
This manuscript was not financed or supported by any institution.
Conflicts of interest/Competing interests
The author declares that they are not aware of competing financial interests or personal relationships that may have influenced the work reported in this article.The author further states that this paper comprises a review article that, for its compilation, it did not require research involving human participants and/or animals.
Availability of data and material
‘Not applicable' for that section.
Code availability
‘Not applicable' for that section.
Authors' contributions
As there is only one author of the manuscript, his contribution was integral and total from its conception until its completion.
Consent to participate
The author of the manuscript is unique and the conception of the article was spontaneous.
Consent for publication
I, as the sole author of the manuscript, authorize the publication of the manuscript if it is accepted.
Ethics approval
‘Not applicable' for that section.
Compliance with ethical standards
The author declares that there are no competing in-terests (financial or non-financial) related to the content of the specific article. The author further states that this paper comprises a review article and that, for its compilation, it did not require research involving human participants and/or animals.
References
- Alaofi A.L., Shahid M. Mutations of SARS-CoV-2 RBD may alter its molecular structure to improve its infection efficiency. Biomolecules. 2021;11(9):1273. doi: 10.3390/biom11091273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O'Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., 2nd, Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H., mRNA-1273 Study Group Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto G., Mohsen M.O., Zinkhan S., Liu X., Vogel M., Bachmann M.F. In vitro data suggest that Indian delta variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2021 doi: 10.1111/all.15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto G., Mohsen M.O., Zinkhan S., Liu X., Vogel M., Bachmann M.F. In vitro data suggest that Indian variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2021 doi: 10.1111/all.15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral P., Bhattarai N., Hossen M.L., Stebliankin V., Gerstman B.S., Narasimhan G., Chapagain P.P. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem. Biophys. Res. Commun. 2021;574:14–19. doi: 10.1016/j.bbrc.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020:13525. doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V., Sousa C., Menezes L., Gonçalves A.M., Picão M., Almeida J.P., Vieita M., Santos R., Silva A.R., Costa M., Carneiro L., Isidro J., Duarte S., Vieira L., Guiomar R., Silva S., Nunes B., Gomes J.P. Tracking SARS-CoV-2 VOC 202012/01 (lineage B.1.1.7) dissemination in Portugal: insights fromnationwide RT-PCR Spike gene drop out data. Virological. Available. 2021. https://virological.org/t/tracking-sars-cov-2-voc-202012-01-lineage-b-1-1-7-dissemination-in-portugal-insights-from-nationwide-rt-pcr-spike-gene-drop-out-data/600 [DOI] [PMC free article] [PubMed]
- Brouwer P.J., Caniels T.G., van der Straten K. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;80 doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., Manuli E.R., Thézé J., Almeida L., Menezes M.T., et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369(6508):1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Science brief: omicron (B.1.1.529) variant. 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html
- CDC 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fmore%2Fscience-and-research%2Fscientific-brief-emerging-variants.html
- Cele S., Gazy I., Jackson L., Hwa S.H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., Karim F., Ganga Y., Khan K., Bernstein M., Balazs A.B., Gosnell B.I., Hanekom W., Moosa M.S. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593(7857):142–146. doi: 10.1038/s41586-021-03471-w. Network for Genomic Surveillance in South Africa; COMMIT-KZN Team, Lessells RJ, de Oliveira T, Sigal A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021 doi: 10.1136/bmj.n579. 372:n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 322 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. 323 Epub 2020 Jul 23. PMID: 32710986; PMCID: PMC7375973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432 doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Wei G.W. 2021. Review of the Mechanisms of SARS-CoV-2 Evolution and Transmission. ArXiv [Preprint]. 2021 arXiv:2109.08148v1. [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., Team N. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;84(369):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P., Caraffa A., Gallenga C.E., Kritas S.K., Frydas I., Younes A., Di Emidio P., Tetè G., Pregliasco F., Ronconi G. The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem. J. Biol. Regul. Homeost. Agents. 2021;35(1):1–4. doi: 10.23812/21-3-E. [DOI] [PubMed] [Google Scholar]
- Davies N.G., Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;3055:1–16. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv [Preprint] 2021 doi: 10.1101/2021.03.07.21252647. 2021.03.07.21252647. [DOI] [Google Scholar]
- Di Giacomo S., Mercatelli D., Rakhimov A., Giorgi F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021;93(9):5638–5643. doi: 10.1002/jmv.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M., Chen R., Xie X., Case J., Zhang X., VanBlargan L., Liu Y., Liu J., Errico J., Winkler E., Suryadevara N., Tahan S., Turner J., Kim W., Schmitz A., Thapa M., Wang D., Boon A., Pinto D., Presti R., O'Halloran J., Kim A., Deepak P., Fremont D., Corti D., Virgin H., Crowe J., Droit L., Ellebedy A., Shi P.Y., Gilchuk P. 2021. SARS-CoV-2 Variants Show Resistance to Neutralization by Many Monoclonal and Serum-Derived Polyclonal Antibodies. Res Sq [Preprint]rs.3.rs-228079. Update in: Nat Med. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicken S.J., Murray M.J., Thorne L.G., Reuschl A.K., Forrest C., Ganeshalingham M., Muir L., Kalemera M.D., Palor M., McCoy L.E., Jolly C., Towers G.J., Reeves M.B., Grove J. 2021. Characterisation of B.1.1.7 and Pangolin Coronavirus Spike Provides Insights on the Evolutionary Trajectory of SARS-CoV-2. bioRxiv. [DOI] [Google Scholar]
- Edara V.V., Norwood C., Floyd K., Lai L., Davis-Gardner M.E., Hudson W.H., Mantus G., Nyhoff L.E., Adelman M.W., Fineman R., Patel S., Byram R., Gomes D.N., Michael G., Abdullahi H., Beydoun N., Panganiban B., McNair N., Hellmeister K., Pitts J., Winters J., Kleinhenz J., Usher J., O'Keefe J.B., Piantadosi A., Waggoner J.J., Babiker A., Stephens D.S., Anderson E.J., Edupuganti S., Rouphael N., Ahmed R., Wrammert J., Suthar M.S. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 2021;29(4):516–521. doi: 10.1016/j.chom.2021.03.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira I.A., Kemp S.A., Datir R., Saito A., Meng B., Rakshit P., Takaori-Kondo A., Kosugi Y., Uriu K., Kimura I., Shirakawa K., Abdullahi A., Agarwal A., Ozono S., Tokunaga K., Sato K., Gupta R.K., CITIID-NIHR BioResource COVID-19 Collaboration, Indian SARS-CoV-2 Genomics Consortium Genotype to phenotype Japan (G2P-Japan) consortium. SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J. Infect. Dis. 2021;224(6):989–994. doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., Fujimoto T., Kuroda M., Wakita T., Ohmagari N. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg. Infect. Dis. 2021;27(4) doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks Jt MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong Gl Biggerstaff M., Dugan V.G. Emergence of SARS-CoV-2 B.1.1.7lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:95e99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidari A., Sabbatini S., Bastianelli S., Pierucci S., Busti C., Monari C., LucianiPasqua B., Dragoni F., Schiaroli E., Zazzi M., Francisci D. Cross-neutralization of SARS-CoV-2 B.1.1.7 and P.1 variants in vaccinated, convalescent and P.1 infected. J. Infect. 2021;S0163–4453(21) doi: 10.1016/j.jinf.2021.07.019. 00362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti M., Benedetti F., Campisi G., Ciccozzi A., Fabris S., Ceccarelli G., Tambone V., Caruso A., Angeletti S., Zella D., Ciccozzi M. Evolution patterns of SARS-CoV-2: snapshot on its genome variants. Biochem. Biophys. Res. Commun. 2021;538:88–91. doi: 10.1016/j.bbrc.2020.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S.M., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., Stalls V., Kopp M.F., Henderson R., Edwards R.J. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34:108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476. doi: 10.1016/j.chom.2021.02.003. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves D.C., Rowland-Jones S.L., Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem. Biophys. Res. Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D., Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P., Graham B., Mascola J., Chang J., Yin M., Sobieszczyk M., Kyratsous C., Shapiro L., Sheng Z., Nair M., Huang Y. 2021. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. Res Sq [Preprint].rs.3.rs-155394. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X., Xu W., Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba G., Sau R., Angioj F., Abbondio M., Rubino S., Uzzau S. A straightforward molecular strategy to retrospectively investigate the spread of SARS-CoV-2 VOC202012/01 B.1.1.7 variant. J. Infect Dev. Ctries. 2021;15(2):242–246. doi: 10.3855/jidc.14972. [DOI] [PubMed] [Google Scholar]
- Istifli E.S., Netz P.A., SihogluTepe A., Sarikurkcu C., Tepe B. Understanding the molecular interaction of SARS-CoV-2 spike mutants with ACE2 (angiotensin converting enzyme 2) J. Biomol. Struct. Dyn. 2021;8:1–12. doi: 10.1080/07391102.2021.1975569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., Garcia-Sastre A., Schotsaert M. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv [Preprint] 2021 doi: 10.1101/2021.01.26.21250543. 2021.01.26.21250543. [DOI] [Google Scholar]
- Jawad B., Adhikari P., Podgornik R., Ching W.Y. Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: combination of molecular dynamics simulation and density functional calculation. J. Chem. Inf. Model. 2021;61(9):4425–4441. doi: 10.1021/acs.jcim.1c00560. [DOI] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Jang U.S., Soh S.M., Lee J.Y., Lee H.R. The impact on infectivity and neutralization efficiency of SARS-CoV-2 lineage B.1.351 pseudovirus. Viruses. 2021;13(4):633. doi: 10.3390/v13040633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirola L. Genetic emergence of B.1.617.2 in COVID-19. New Microbes New Infect. 2021;43:100929. doi: 10.1016/j.nmni.2021.100929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K., Toyoshima Y., Nemoto K., Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet.65(7) 2020:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Singh J., Hasnain S.E., Sundar D. Possible link between higher transmissibility of Alpha, kappa and delta variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. Int. J. Mol. Sci. 2021;22(17):9131. doi: 10.3390/ijms22179131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lazarevic I., Pravica V., Miljanovic D., Cupic M. Immune evasion of SARS-CoV-2 emerging variants: what HaveWe learnt SoFar? Viruses. 2021;13:1192. doi: 10.3390/v13071192. v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I., Wang L.F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G., Tan Y.J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1):2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. Erratum in: Euro Surveill. 2021 Jan;26(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;3(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. 182. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H., Baek A., Kim J., Kim M.S., Liu J., Nam K.Y., Yoon J., No K.T. Hot spot profiles of SARS-CoV-2 and human ACE2 receptor protein protein interaction obtained by density functional tight binding fragment molecular orbital method. Sci. Rep. 2020;10(1):16862. doi: 10.1038/s41598-020-73820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Van Blargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477–488. doi: 10.1016/j.chom.2021.01.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Johnson B.A., Xia H., Ku Z., Schindewolf C., Widen S.G., An Z., Weaver S.C., Menachery V.D., Xie X., Shi P.Y. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv [Preprint] 2021 doi: 10.1101/2021.08.12.456173. 2021.08.12.456173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., Duyvesteyn H.M.E., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Johnson S.A., Ritter T.G., Mason C., Costa Clemens S.A., Gomes Naveca F., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Temperton N., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Malik T., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Baillie V., Serafin N., Ditse Z., Da Silva K., Paterson N.G., Williams M.A., Hall D.R., Madhi S., Nunes M.C., Goulder P., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220–4236. doi: 10.1016/j.cell.2021.06.020. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Chu A.W., Zhang R.R., Chan W.M., Ip J.D., Tsoi H.W., Chen L.L., Cai J.P., Lung D.C., Tam A.R., Yau Y.S., Kwan M.Y., To W.K., Tsang O.T., Lee L.L., Yi H., Ip T.C., Poon R.W., Siu G.K., Mok B.W., Cheng V.C., Chan K.H., Yuen K.Y., Hung I.F., To K.K. The impact of spike N501Y mutation on neutralizing activity and RBD binding of SARS-CoV-2 convalescent serum. EBioMedicine. 2021;71:103544. doi: 10.1016/j.ebiom.2021.103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B., Tang T., Daniel S., Jaimes J.A., Whittaker G.R. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: role of the P681H mutation. bioRxiv [Preprint] 2021 doi: 10.1101/2021.04.06.438731. 2021.04.06.438731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Klein J., Sundaram M.E., Liu F., Wong P., Silva J., Mao T., Oh J.E., Mohanty S., Huang J., Tokuyama M., Lu P., Venkataraman A., Park A., Israelow B., Vogels C.B.F., Muenker M.C., Chang C.H., Casanovas-Massana A., Moore A.J., Zell J., Fournier J.B., Impact Research Team Yale, Wyllie A.L., Campbell M., Lee A.I., Chun H.J., Grubaugh N.D., Schulz W.L., Farhadian S., Dela Cruz C., Ring A.M., Shaw A.C., Wisnewski A.V., Yildirim I., Ko A.I., Omer S.B., Iwasaki A. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med. 2021;27(7):1178–1186. doi: 10.1038/s41591-021-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley A.J., Kuiper M.J., Durr P.A., Bruce M.P., Barr J., Todd S., Au G.G., Blasdell K., Tachedjian M., Lowther S., Marsh G.A., Edwards S., Poole T., Layton R., Riddell S.J., Drew T.W., Druce J.D., Smith T.R.F., Broderick K.E., Vasan S.S. Experimental and in silico evidence suggests vaccines are unlikely to be affected by D614G mutation in SARS-CoV-2 spike protein. NPJ Vaccines. 2020;5(1):96. doi: 10.1038/s41541-020-00246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor O., Mandelboim M., Fleishon S., Bucris E., Bar-Ilan D., Linial M., Nemet I., Kliker L., Lustig Y., Israel National Consortium For Sars-CoV-Sequencing, Mendelson ES, Zuckerman NS The rise and fall of a local SARS-CoV-2 variant with the spike protein mutation L452R. Vaccines (Basel) 2021;9(8):937. doi: 10.3390/vaccines9080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., Yue Y., Shimizu R., Ito J., Torii S., Yonekawa A., Shimono N., Nagasaki Y., Minami R., Toya T., Sekiya N., Fukuhara T., Matsuura Y., Schreiber G., Genotype to Phenotype Japan (G2P-Japan) Consortium, Ikeda T, Nakagawa S, Ueno T, Sato K SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7):1124–1136. doi: 10.1016/j.chom.2021.06.006. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A.M., Bianco C., Denu L., Ahmed A., Coffin S.E., Neide B., Everett J., Reddy S., Rabut E., Deseignora J., Feldman M.D., Rodino K.G., Bushman F., Harris R.M., Chang Mell J., Planet P.J. Comparative analysis of emerging B.1.1.7+E484K SARS-CoV-2 isolates. Open Forum Infect. Dis. 2021;8(7) doi: 10.1093/ofid/ofab300. ofab300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö Dormitzer PR., Şahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371(6534):1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella S., Ciccozzi M., Zella D., Bianchi M., Benetti F., Benvenuto D., Broccolo F., Cauda R., Caruso A., Angeletti S., Giovanetti M., Cassone A. SARS-CoV-2 B.1.617 Indian variants: are electrostatic potential changes responsible for a higher transmission rate? J. Med. Virol. 2021 doi: 10.1002/jmv.27210. Epub ahead of print. PMID: 34260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, Thomas P., Goldhill Daniel H., Zhou Jie, Baillon Laury, Frise Rebecca, Swann Olivia C., Kugathasan Ruthira, et al. The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. Cold Spring Harbor Lab. 2020 doi: 10.1101/2020.09.30.318311. [DOI] [Google Scholar]
- Pearson C., Russell T.W., Davies N.G., Kuckarski A.J., CMMID COVID-19 working group . 2021. Estimates of Severity and Transmissibility of Novel South Africa SARS-CoV-2 Variant 501Y.V2 Preprint. Retrieved from: pdf (cmmid.github.io)pdf icon. [Google Scholar]
- Peters M.H., Bastidas O., Kokron D.S., Henze C.E. Transformations, lineage comparisons, and analysis of down-to-up protomer states of variants of the SARS-CoV-2 prefusion spike protein, including the UK variant B.1.1.7. Microbiol. Spectr. 2021;9(1) doi: 10.1128/Spectrum.00030-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Polgár L. in Mechanisms of Protease Action Ch. 1989;2:43–86. CRC press, 1989. [Google Scholar]
- Public Health England . Public Health England; London, United Kingdom: 2021. Investigation of Novel SARS-CoV-2 Variant: Variant of Concern 202012/01, Technical Briefing 3.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/950823/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3_-_England.pdf 2020. [Google Scholar]
- Public Health England Investigation of novel SARS-CoV-2 variant. 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1036501/Technical_Briefing_29_published_26_November_2021.pdf
- Public Health England SARS-CoV-2 variants of concern and variants under investigation in England. 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1040076/Technical_Briefing_31.pdf
- Queirós-Reis L., Gomes da Silva P., Gonçalves J., Brancale A., Bassetto M., Mesquita J.R. SARS-CoV-2 Virus−Host interaction: currently available structures and implications of variant emergence on infectivity and immune response. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910836. 10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., et al. 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/
- Rodríguez-Maldonado A.P., Vázquez-Pérez J.A., Cedro-Tanda A., Taboada B., Boukadida C., Wong-Arámbula C., Nuñez-García T.E., Cruz-Ortiz N., Barrera-Badillo G., Hernández-Rivas L., López-Martínez I., Mendoza-Vargas A., Reyes-Grajeda J.P., Alcaraz N., Peñaloza-Figueroa F., Gonzalez-Barrera D., Rangel-DeLeon D., Herrera-Montalvo L.A., Mejía-Nepomuceno F., Hernández-Terán A., Mújica-Sánchez M., Becerril-Vargas E., Martínez-Orozco J.A., Pérez-Padilla R., Salas-Hernández J., Sanchez-Flores A., Isa P., Matías-Florentino M., Ávila-Ríos S., Muñoz-Medina J.E., Grajales-Muñiz C., Salas-Lais A.G., Santos Coy-Arechavaleta A., Hidalgo-Miranda A., Arias C.F., Ramírez-González J.E. Emergenceand spread ofthepotentialvariantofinterest (VOI) B.1.1.519 of SARS-CoV-2 predominantlypresent in Mexico. Arch. Virol. 2021 doi: 10.1007/s00705-021-05208-6. l1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Nasser H., Uriu K., Kosugi Y., Irie T., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., Ozono S., Tokunaga K., Butlertanaka E.P., Tanaka Y.L., Shimizu R., Shimizu K., Fukuhara T., Kawabata R., Sakaguchi T., Yoshida I., Asakura H., Nagashima M., Yoshimura K., Kazuma Y., Nomura R., Horisawa Y., Takaori-Kondo A., Nakagawa S., Ikeda T., Sato K. 2021. SARS-CoV-2 Spike P681R Mutation Enhances and Accelerates Viral Fusion. bioRxiv doi:10.1101/2021.06.17.448820:2021.06.17.448820. [Google Scholar]
- Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2022;12:830527. doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzamani K., Mahmoudian F., Ahangarzadeh S., Ranjbar M.M., Beikmohammadi L., Bahrami S., Mohammadi E., Esfandyari S., Alibakhshi A., Javanmard S.H. Vaccine design and delivery approaches for COVID-19. Int. Immunopharm. 2021;23(100):108086. doi: 10.1016/j.intimp.2021.108086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., Gnanakaran S., Hengartner N., Pajon R., Smith G., Glenn G.M., Korber B., Montefiori D.C. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529–539. doi: 10.1016/j.chom.2021.03.002. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiehzadegan S., Alaghemand N., Fox M., Venketaraman V. Analysis of the delta variant B.1.617.2 COVID-19. Clin. Pract. 2021;11(4):778–784. doi: 10.3390/clinpract11040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira M.M., Moreira G.M., Mendonça M. DNA vaccines against COVID-19: perspectives and challenges. Life Sci. 2021;267:118919. doi: 10.1016/j.lfs.2020.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Pandit P., McArthur A.G., Banerjee A., Mossman K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021;18(1):166. doi: 10.1186/s12985-021-01633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom JD.Deep Mutational Scanning of SARS-CoV-2 Receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell182(5) 2020:1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., D costa B.M., Zhou H., Vaill A., Kazmierski W., Landau N.R. Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. bioRxiv [Preprint] 2021 doi: 10.1101/2021.02.18.431897. 2021.02.18.431897. [DOI] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., Landau N.R. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv [Preprint] 2021 doi: 10.1101/2021.02.05.430003. 2021.02.05.430003. [DOI] [Google Scholar]
- Tchesnokova V., Kulakesara H., Larson L., Bowers V., Rechkina E., Kisiela D., Sledneva Y., Choudhury D., Maslova I., Deng K., Kutumbaka K., Geng H., Fowler C., Greene D., Ralston J., Samadpour M., Sokurenko E. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv [Preprint] 2021 doi: 10.1101/2021.02.22.432189. 2021.02.22.432189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel N., Mailhot O., Najmanovich R.J. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 Spike protein variants. PLoS Comput. Biol. 2021;17(8) doi: 10.1371/journal.pcbi.1009286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10 doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan G. Coronavirus variants are spreading in India — what scientists know so far. Nature. 2021;593:321–322. doi: 10.1038/d41586-021-01274-7. [DOI] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., COG-UK Consortium, Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.B., Liang Y., Jin Y.Q., Zhang J., Su J.G., Li Q.M. E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: binding free energy calculation studies. J. Mol. Graph. Model. 2021;17(109):108035. doi: 10.1016/j.jmgm.2021.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751. doi: 10.1016/j.chom.2021.04.007. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen J., Hozumi Y., Yin C., Wei G.W. Emerging vaccine-breakthrough SARS-CoV-2 variants. ACS Infect. Dis. 2022;8(3):546–556. doi: 10.1021/acsinfecdis.1c00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J.N., Moore P.L. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-X. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 42. World Health Organization; 2021. COVID-19 Weekly Epidemiological Update.https://apps.who.int/iris/handle/10665/341622 1 June 2021. [Google Scholar]
- World Health Organization 2021. https://cdn.who.int/media/docs/default-source/searo/whe/searo-technical-brief-enhacing-readiness-on-omicron.pdf?sfvrsn=6457b0d7_15
- World Health Organization . World Health Organization; Geneva: 2021. COVID-19 Weekly Epidemiological Update [Internet]https://www.who.int/docs/default-source/coronaviruse/situationreports/20210511_weekly_epi_update_39.pdf?sfvrsn=b66ba70d_5&download=true [updated 2021 May 11; cited 2021 May 12]. Available from. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;13(367):1260–1263. doi: 10.1126/science.abb2507. 6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., Moliva J.I., Sullivan N.J., Graham B.S., Carfi A., Corbett K.S., Seder R.A., Edwards D.K. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384(15):1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang Y., Qu Y., Zhang C., Liu X.W., Zhao M., Mu Y., Li W. Key residues of the receptor binding domain in the spike protein of SARS-CoV-2 mediating the interactions with ACE2: a molecular dynamics study. Nanoscale. 2021;13(20):9364–9370. doi: 10.1039/d1nr01672e. [DOI] [PubMed] [Google Scholar]
- Yarmarkovich M., Warrington J.M., Farrel A., Maris J.M. Identification of SARS-CoV-2 vaccine epitopes predicted to induce long-term population-scale immunity. Cell Rep. Med. 2020;1(3):100036. doi: 10.1016/j.xcrm.2020.100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazhini A., Sidhanta D.S.P., Srinivasan N. D614G substitution at the hinge region enhances the stability of trimeric SARS-CoV-2 spike protein. Bioinformation. 2021;17(3):439–445. doi: 10.6026/97320630017439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., Burton D.R., Reincke S.M., Prüss H., Kreye J., Nemazee D., Ward A.B., Wilson I.A. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373(6556):818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A., Dudkina N.V., Shen K., Luban J. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751. doi: 10.1016/j.cell.2020.09.032. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Ran J., Han L. Exploring the interaction between E484K and N501Y substitutions of SARS-CoV-2 in shaping the transmission advantage of COVID-19 in Brazil: a modeling study. Am. J. Trop. Med. Hyg. 2021 doi: 10.4269/ajtmh.21-0412. tpmd210412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. China novel coronavirus investigating and research team. N. Engl. J. Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Yunkai, FeiFeng, Hu Gaowei, Wang Yuyan, Yu Yin, Zhu Yuanfei, Xu Wei, et al. The S1/S2 boundary of SARS-CoV-2 spike protein modulates cell entry pathways and transmission. Cold Spring Harbor Lab. 2020 doi: 10.1101/2020.08.25.266775. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
‘Not applicable' for that section.