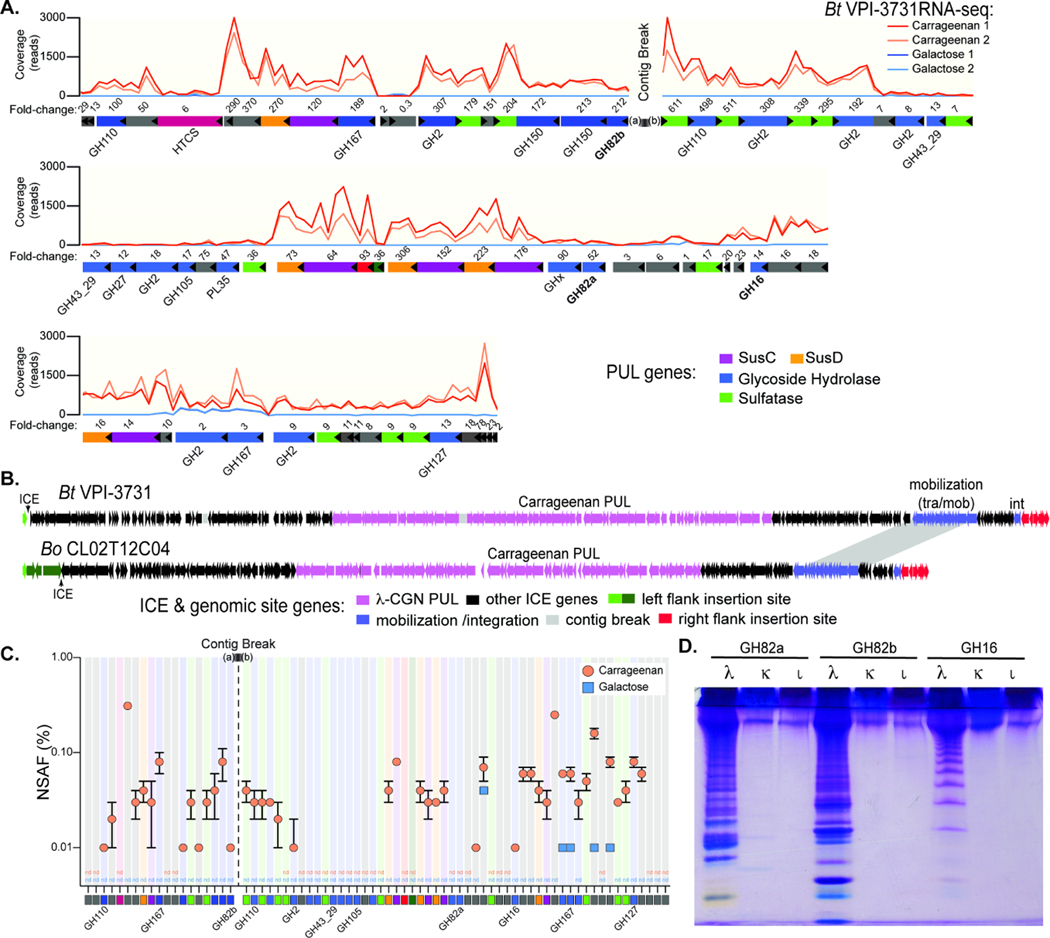
Figure 2. Bacteroides genes and enzyme involved in carrageenan degradation.
(A) Transcriptional responses of genes within the 130.5kbp candidate λ-CGN PUL in Bt3731 during growth on λ-CGN compared to a galactose reference. Numbers above genes refer to fold change induction by CGN relative to galactose (Table S2A). Two replicate experiments were conducted for each treatment condition. (B) A comparison of the surrounding Bt3731 and Bo12C04 ICE architecture showing the genes at the flanking chromosomal insertion gene in green and red (see Figure S2B for higher resolution view of insertion site). (C) Relative abundances (in % of normalized spectral abundance factor, NSAF) of PUL-encoded proteins detected in the total protein fraction of cultures grown on carrageenan (orange) and galactose (blue). Error bars are the standard error of the mean (SEM). Proteins that could be detected in at least two out of three independent biological replicates of each substrate condition are shown and color coded squares are at bottom to match genetic architecture in (A). Three replicate experiments were conducted for each treatment condition. (D) C-PAGE gel showing digestion of three different CGN isomer types (λ, lambda; κ, kappa; Ɩ, iota) with recombinant GH16 and GH82 enzymes that are highly expressed within the Bt3731 PUL. Shorter CGN oligosaccharides migrate toward the bottom of the gel.