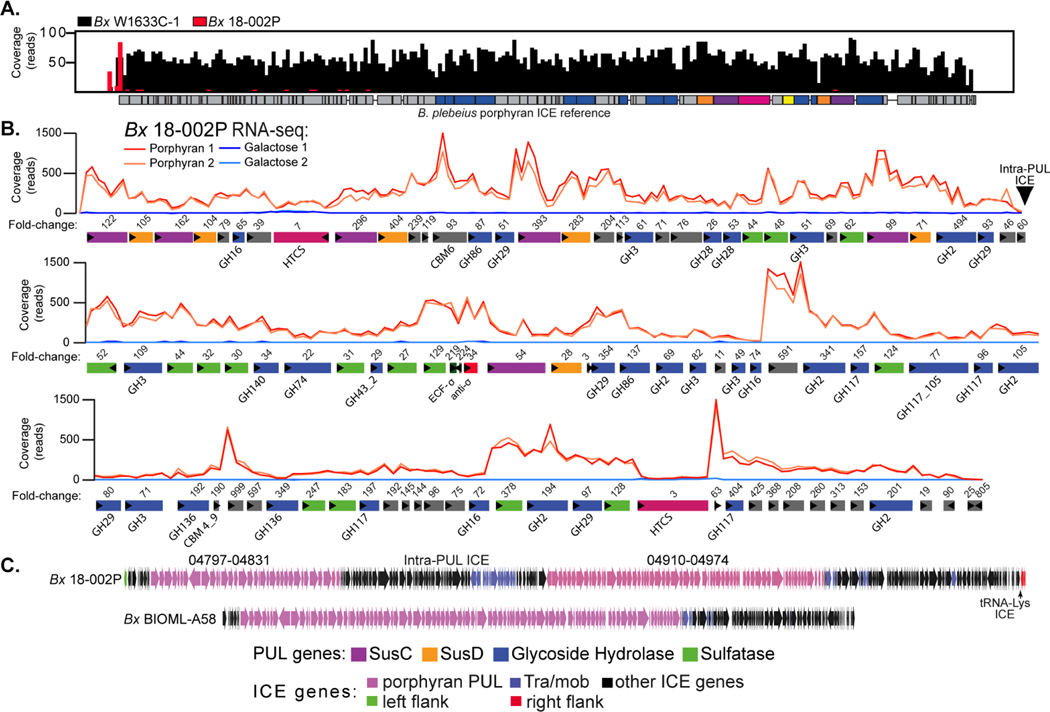
Figure 4. Identification of a second mobile element and PUL that is associated with porphyran degradation by human gut Bacteroides.
(A) Histogram of reference-guided mapping results to the B. plebeius porphyran ICE for two different strains: BxW1633C−1 (black) and Bx18−002P (red). BxW1633C−1 has full coverage across the entire Bp ICE, while sequences for Bx18−002P do not align to the known B. plebeius genes, except for a small region at the far left. (B) RNA-seq based transcriptional responses of genes within a 168.6kbp PUL in Bx18−002P during growth on porphyran compared to galactose. Numbers of above genes refer to fold change induction by prophyran relative to galactose (Table S2B). Two replicate experiments were conducted for each treatment condition. (C) Lower resolution comparison of the ICEs between the Bx18−002P and BxBIOML-A58 strains, highlighting the presence of extra genes, possibly the result of an additional ICE insertion into the middle of the Bx18−002P PUL without disrupting its function.