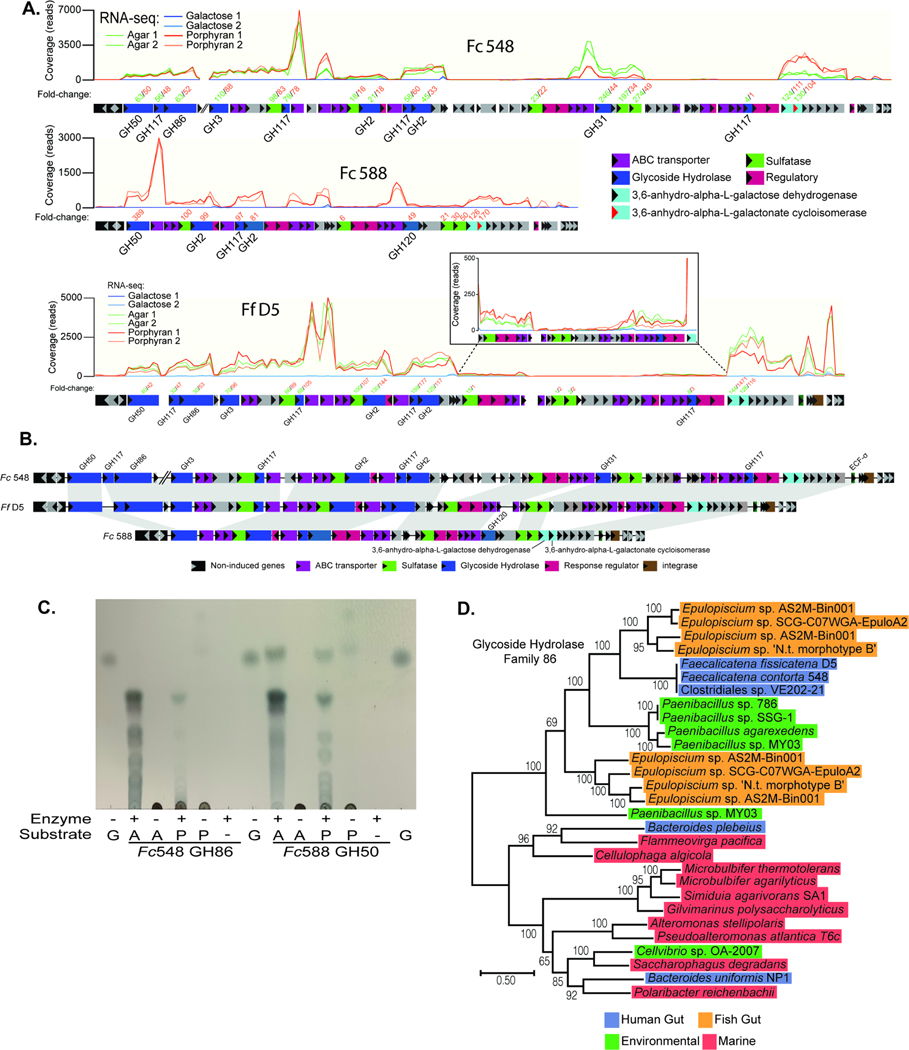
Figure 5. Identification of genes in human gut Firmicutes associated with agarose and porphyran utilization.
(A) RNA-seq based transcriptional responses of the homologous loci in Fc548, Fc588 and Ff D5 during growth on agarose and/or porphyran compared to galactose. The inset box shows a closer view of a section of genes that are induced less compared to their surrounding neighbors, but still show a positive response compared to galactose. Numbers of above genes refer to fold change induction by agarose or porphyran (according to color code when applicable) relative to galactose (Tables S2C-E). Two replicate experiments were conducted for each treatment condition. (B) A genomic comparison of the three Faecalicatena spp. locus maps showing synteny and variation among the genes responsible for degrading agarose and/or porphyran. Note that the first three enzymes in Fc548 are separated genomically from the rest of the genes in the loci by 110 genes (∼87.9 kbp). (C) Thin-layer chromatography of Fc548 GH86 and Fc588 GH50 enzymes on high molecular weight agarose (A) and porphyran (P) with galactose (G) serving as a monosaccharide standard. (D) Phylogenetic position of Faecalicatena GH86 enzymes compared to other gut and environmental enzymes in this family, highlighting the similarity to enzymes found in Epulopiscium and Paenibacillus spp. Clostridiales sp. VE-202–2 is a Faecalicatena contorta isolate from a Japanese adult that has a copy of this locus and likely degrades these polysaccharides.