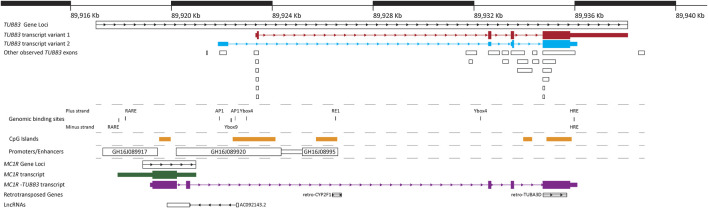
FIGURE 2.
Structure of the human TUBB3 loci. A map of the human TUBB3 loci showing the main two transcripts of TUBB3 as well as several other genomic structures and DNA binding sites in the region. Gene loci’s (TUBB3, MC1R, retro-CYP2F1 and retro-TUBA3D) are represented by boxes with internal arrows showing sequence direction. Individual RNA transcripts are represented by a combination of thin and thick boxes for exons, and arrows for introns; thin boxes represent untranslated regions (5′ and 3′ UTRs), while thick boxes represent translated regions. TUBB3 transcript variants 1 and 2 are show in red and blue respectively, with addition observed TUBB3 exons presented in white boxes; additional information of TUBB3 transcripts is shown in Table 1. MC1R transcript is shown in green, the MC1R-TUBB3 chimera transcript is shown in purple, and the lncRNA AC092143.2 is shown in white. Known genomic binding sites are represented with black lines, with thickness corresponding to size; additional information on known genomic binding sites is shown in Table 2 . CpG islands presented within the TUBB3 loci are represented by orange rectangles. Promoters/Enhancers are shown as white boxes with their name. Locations of TUBB3, MC1R and MC1R-TUBB3 transcripts, and CpG islands was extracted from the Ensembl database (Cunningham et al., 2018). Promoters/Enhancers sourced from Genehancer (Fishilevich et al., 2017). Location of retro-transposed genes and lncRNAs sourced from the UCSC genome browser (Haeussler et al., 2019). Positions based on human genome GCRh38/hg38 assembly.