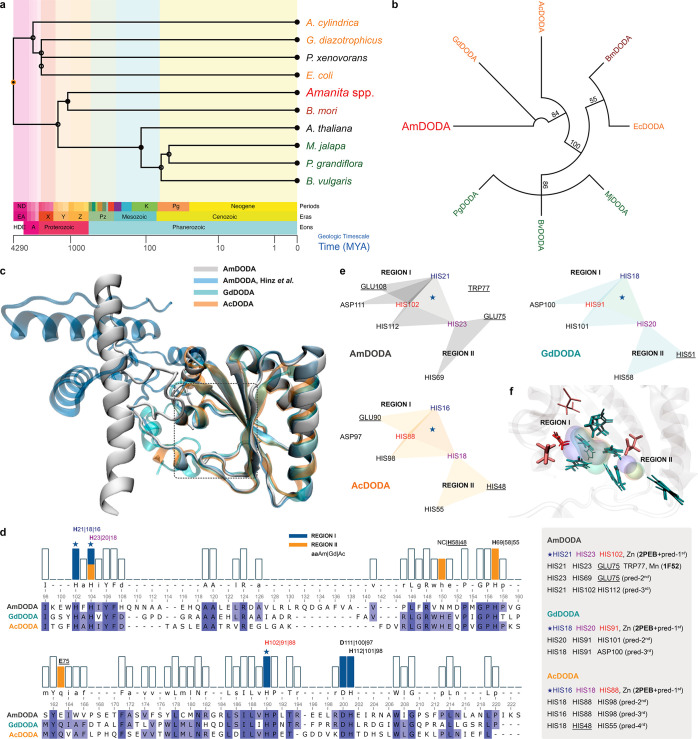
Figure 4.
Phylogenetic relations and structural models of DODAs. (a) Divergence times for selected species producing dioxygenases. (b) Neighbor-Joining consensus tree inferred from protein sequences of functionally characterized l-DOPA extradiol dioxygenases (DODAs). Different groups of organisms are presented in the following colors: fungi (red), bacteria (orange), insects (brown), and plants (green). Branch support values >50% are indicated. Except for AmDODA, whose amino acid sequence was generated from the translation of the 558-bp CDS, protein sequences were obtained from the Uniprot database. (c) Structural alignment of AmDODA, GdDODA, and AcDODA. Enzymes were treated as monomeric units for simplicity, and the cutoff distance for the α-Cs was set to 140 pm (picometer). (d) Sequence alignment according to the structural alignment, and the results of the metal site prediction and homology search. The two regions of the putative catalytic pocket were assigned as I (blue) and II (orange). The star indicates the highest score motif for all three sequences. (e) Amino acid residues that are involved in complexation. Triangles show triads of amino acids that were predicted to bind metal cations in AmDODA (gray), GdDODA (green), and AcDODA (orange); underlined aa are not conserved in all three enzymes. (f) Amino acids at the putative catalytic pocket of the three DODAs. His (H), Glu (E), and Asp (D) residues are colored in cyan, pink, and red, respectively. Thick residues are from AmDODA. Colored volumes show the most probable region of metal cation binding; AmDODA (blue), GdDODA (green), and AcDODA (orange).