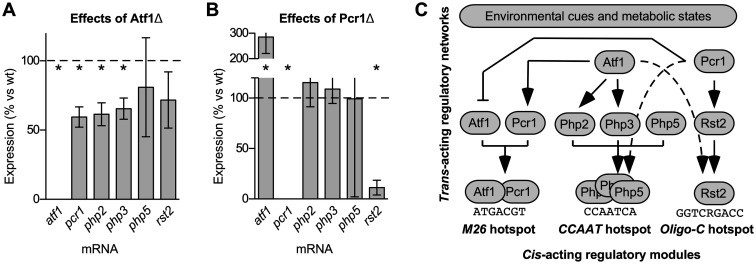
Figure 5.
Hotspot-activating proteins can regulate the expression of other hotspot-activating proteins. Quantitative, reverse-transcription PCR was used to determine the effects of Atf1 and Pcr1 on the expression of the six genes (atf1, pcr1, php2, php3, php5, and rst2) that encode hotspot-binding/activating proteins. Gene expression levels in mutants are plotted as % relative to wild-type for: (A) atf1Δ mutant; (B) pcr1Δ mutant. Data are mean ± SD from four independent biological replicates; differences with P ≤ 0.05 are indicated for mutant vs wild-type (*). (C) Conclusions from this and prior figures. The impacts of environmental conditions on meiotic recombination are mediated via cis-acting regulatory modules (site-specific protein-DNA complexes). Transcriptional regulation of hotspot-binding/activating proteins and cross-talk between regulatory networks each contribute to the control of hotspots. Arrows depict transcription-mediated pathway connections elucidated in this study (solid lines in upper half of panel), as well as connections where hotspot control in trans was also observed, but where there were no significant changes in expression of the genes encoding the respective activator proteins (dotted lines).