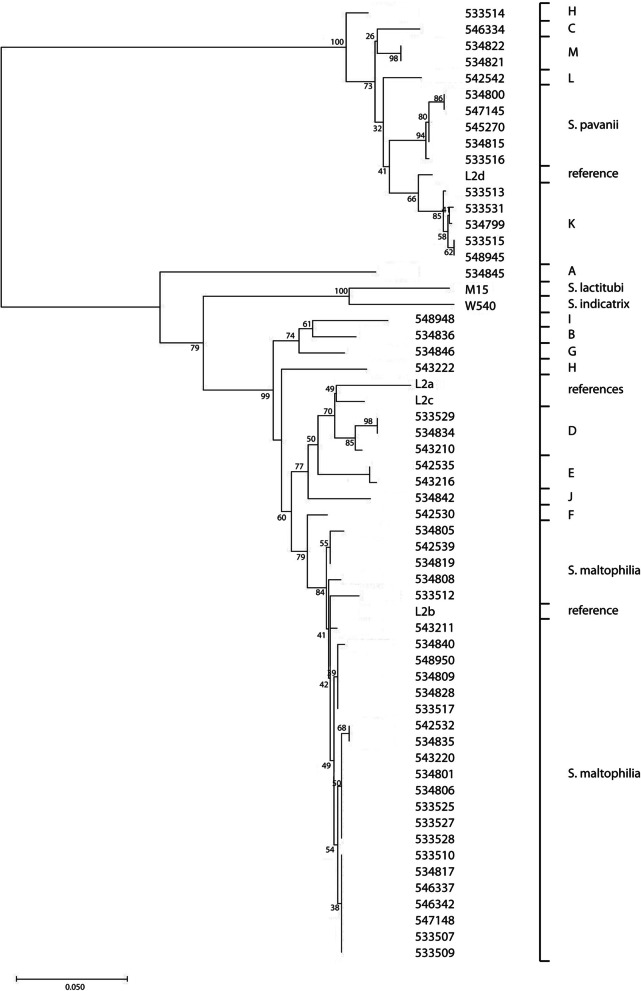
Fig. 5.
The evolutionary history of L2 ß-lactamases was inferred using the Neighbor-Joining method. One amino acid sequence per ST was included. L2a-L2d were used as reference sequences from literature (Walsh et al., 1997). Strains M15 and W540 are the type strains for S. lactitubi and S. indicatrix, respectively. The optimal tree with the sum of branch length = 1.04210294 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1050 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 59 amino acid sequences. There were a total of 305 positions in the final dataset. There were a total of 304 positions in the final dataset. No L2 ß-lactamase sequences for ST89 and ST215 isolates were found