Abstract
Background & Aims
Alpha-1 antitrypsin deficiency is caused by mutations in SERPINA1, most commonly homozygosity for the Pi∗Z variant, and can present as liver disease. While heterozygosity for Pi∗Z (Pi∗MZ) is linked to increased risk of cirrhosis, whether the Pi∗MZ genotype is associated with an increased rate of decompensation among patients who already have compensated cirrhosis is not known.
Methods
This was a retrospective study of Michigan Genomics Initiative participants with baseline compensated cirrhosis. The primary predictors were Pi∗MZ or Pi∗MS genotype (vs. Pi∗MM). The primary outcomes were hepatic decompensation with ascites, hepatic encephalopathy, or variceal bleeding, or the combined endpoint of liver-related death or liver transplant, both modeled with Fine-Gray competing risk models.
Results
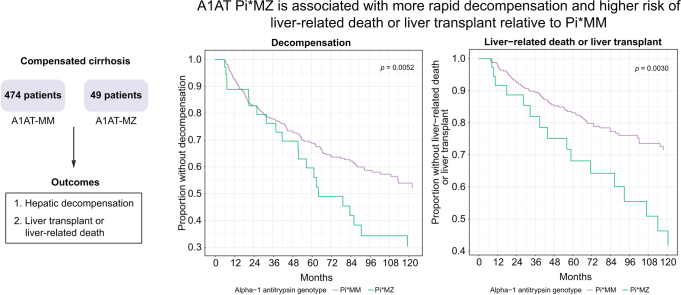
We included 576 patients with baseline compensated cirrhosis who had undergone genotyping, of whom 474 had Pi∗MM, 49 had Pi∗MZ, and 52 had Pi∗MS genotypes. Compared to Pi∗MM genotype, Pi∗MZ was associated with increased rates of hepatic decompensation (hazard ratio 1.81; 95% CI 1.22-2.69; p = 0.003) and liver transplant or liver-related death (hazard ratio 2.07; 95% CI 1.21-3.52; p = 0.078). These associations remained significant after adjustment for severity of underlying liver disease, and were robust across subgroup analyses based on etiology, sex, obesity, and diabetes status. Pi∗MS was not associated with decompensation or death/transplantation.
Conclusions
The SERPINA1 Pi∗MZ genotype is associated with an increased rate of hepatic decompensation and decreased transplant-free survival among patients with baseline compensated cirrhosis.
Lay summary
There is a mutation in the gene SERPINA1 called Pi∗MZ which increases risk of liver scarring (cirrhosis); however, it is not known what effect Pi∗MZ has if someone already has cirrhosis. In this study, we found that people who had cirrhosis and Pi∗MZ developed complications from cirrhosis faster than those who did not have the mutation.
Keywords: Genetic epidemiology, single nucleotide polymorphism, survival analysis, competing risk
Abbreviations: A1AT, alpha-1 antitrypsin; CSPH, clinically significant portal hypertension; HR, hazard ratio; MELD, model for end-stage liver disease; MGI, Michigan Genomics Initiative; NAFLD, non-alcoholic fatty liver disease; VCTE, vibration-controlled transient elastography
Graphical abstract
Highlights
-
•
We evaluated the impact of SERPINA Pi∗MZ in a compensated cirrhosis cohort.
-
•
SERPINA1 Pi∗MZ was associated with an increased risk of decompensation vs. Pi∗MM.
-
•
Pi∗MZ was associated with an increased risk of liver-related death or liver transplant.
-
•
Findings were robust across several sensitivity analyses.
Introduction
Alpha-1 antitrypsin (A1AT) deficiency is estimated to affect around 100,000 people in the United States and usually presents as chronic obstructive pulmonary disease or chronic liver disease, which ranges from asymptomatic liver enzyme elevations to chronic hepatitis and cirrhosis with portal hypertensive complications.1,2 A1AT deficiency is caused by SERPINA1 mutations, the most clinically relevant of which are the Pi∗Z (E366K; rs28929474-T) and Pi∗S (E288V; rs17580-A) alleles, while Pi∗M corresponds to wild-type A1AT. Pi∗ZZ carries the most severe phenotype, is characterized by severe A1AT deficiency (10-20% normal), and is associated with the highest risk of liver and lung disease. Up to 13% of patients with the Pi∗ZZ genotype die of liver disease and 55% of lung disease.3 In contrast, Pi∗MZ, Pi∗SZ, and Pi∗SS have a milder phenotype with higher levels of A1AT (30-60% normal) and lower risk of cirrhosis. Notably, liver disease in A1AT deficiency is not caused by protein deficiency per se, but rather by misfolding of the A1AT protein induced by specific SERPINA1 mutations (most notably Pi∗Z) which results in accumulation in the endoplasmic reticulum and decreased secretion of the protein from hepatocytes.4
While clinically diagnosed A1AT deficiency is relatively uncommon, the Pi∗Z and Pi∗S alleles are relatively common with estimated minor allele frequency of 0.037 and 0.016, respectively, in Caucasians; this discrepancy may be due to low penetrance and disease awareness.2,5 Given this, there are a number of individuals carrying the Pi∗Z or Pi∗S allele who are unaware of this genotype yet may have increased risk of disease, even if penetrance is low.6 One study from a Swedish national registry of 1,595 individuals with the Pi∗ZZ genotype found that 26% had persistently elevated liver enzymes, while 7% developed cirrhosis during follow-up.7 Pi∗ZZ appeared to interact with established risk factors such as viral hepatitis and diabetes as these patients were at higher risk of developing liver disease.7 Another multinational European study found that Pi∗ZZ was associated with an odds ratio of 16.3 for liver stiffness measurement ≥10 kPa on vibration-controlled transient elastography (VCTE) compared to Pi∗Z non-carriers.8 Pi∗ZZ was also associated with an increased risk of severe hepatic steatosis based on controlled attenuation parameter measured by VCTE (odds ratio 2.1 for severe steatosis).8
The associations between Pi∗MZ or Pi∗SZ and liver disease appear to be weaker. One study of the UK Biobank found modestly increased risk of liver fibrosis/cirrhosis with Pi∗MZ (odds ratio 1.7) and Pi∗SZ (odds ratio 3.1) relative to Pi∗MM.9 We also reported a dose-dependent increase in risk of cirrhosis based on Pi∗Z dosage in a UK Biobank and a United States cohort.5 Of note, whether the participants with more advanced liver disease all had other underlying liver diseases was not reported, so it is uncertain whether Pi∗MZ is sufficient to cause cirrhosis on its own. One Austrian study evaluated 596 individuals with Pi∗MM or Pi∗MZ genotypes and cirrhosis, and found that Pi∗MZ carriers were at an increased risk of ascites and hepatic encephalopathy and a higher model for end-stage liver disease (MELD) score, with no significant difference in transplant-free survival.10 Taken together, the existing data suggest that Pi∗ZZ is the most deleterious variant, followed by Pi∗SZ, then Pi∗MZ. Pi∗SS and Pi∗MS have not been consistently shown to increase risk of liver disease.9
The literature to date has been primarily based in Europe, with relatively little literature from the United States. Further, most studies determined presence or absence of liver disease but not rate of disease progression based on SERPINA1 genotype. There is also minimal literature on the impact of A1AT deficiency in patients who already have baseline compensated cirrhosis. In this study, we sought to characterize the impact of the A1AT Pi∗MZ and Pi∗MS variants on the rate of hepatic decompensation using time-to-event survival analyses in patients with established cirrhosis.
Patients and methods
Ethics
Michigan Genomics Initiative (MGI) participants provided written informed consent approved by the Institutional Review Board of the University of Michigan (Ann Arbor, MI), which also approved all research conducted in this study.
Cohort and genotyping
MGI is a prospective cohort with ongoing enrollment and at time of analysis included >70,000 individuals. MGI recruits primarily from patients undergoing elective surgery at Michigan Medicine (Ann Arbor, MI) as well as from other selected outpatient populations such as patients with diabetes followed in endocrinology clinics. Participants underwent genotyping of whole blood on an Illumina HumanCoreExome v.12.1 array, a combined genome-wide association study and exome array consisting of >500,000 single nucleotide polymorphisms.11 Imputation was performed based on the Haplotype Reference Consortium (release 1 for chromosomes 1-22 and 1.1 for X).12
A1AT Pi∗Z was defined as rs28929474-T and Pi∗S by rs17580-A, while Pi∗M was defined by the absence of either rs28929474-T or rs17580-A. Other SERPINA1 genotypes were not evaluated in this analysis.
Liver phenotypes
We screened for cirrhosis based on a combination of diagnostic codes for cirrhosis and portal hypertension (Table S1), endoscopic evidence of varices, VCTE liver stiffness measurement >16 kPa, imaging evidence of cirrhosis, or liver biopsy reports showing cirrhosis. We obtained data on presence of varices with an automated algorithm that screened for the terms “varix”, “varic”, “GOV1”, “GOV2”, “IVG1”, and “IGV2” in upper endoscopy reports. Reports that included any of these terms without a negative term (e.g., “no”) or an assessment term (e.g., “rule out”) in the same sentence were flagged as representing cirrhosis. VCTE reports at our institution are standardized to report liver stiffness measurement in a designated field, and these values were extracted using an automated algorithm, with a cut-off of 16 kPa for cirrhosis chosen to maximize specificity.13 Imaging and liver biopsy reports were screened for evidence of cirrhosis with an algorithm near-identical to the one we previously reported except that instead of assigning patients as cases vs. controls, herein, we were only interested in identifying cases.5
Diagnoses of cirrhosis, varices, and hepatic decompensation were then verified manually (DAB, IJM, JVD, MJM) and the dates of diagnosis of cirrhosis and/or complications were recorded. Cirrhosis was defined based on compatible imaging, biopsy, or evidence of portal hypertension without an alternative explanation. Hepatic decompensation was defined as (1) ascites severe enough to require treatment (diuretics, paracentesis, or transjugular intrahepatic portosystemic shunt), (2) hepatic encephalopathy, or (3) variceal hemorrhage. The automated algorithms for analyzing reports for VCTE, imaging, liver biopsy, or upper endoscopy all had specificity >98% for their respective criteria for cirrhosis (n = 72, 505, 127, and 218, respectively). The accuracies of diagnostic codes for cirrhosis were previously reported to be high with the exception of codes for ascites.14
Statistics
Quantitative variables were reported as median (interquartile range). Categorical variables were reported as percentages. Three-way comparisons of continuous variables were performed with Kruskal-Wallis statistics, and categorical variables were compared with a chi-squared statistic.
In survival analyses, the primary predictor was Pi∗MZ or Pi∗MS genotype (vs. Pi∗MM). The primary outcomes were hepatic decompensation and a combined outcome of liver transplant or liver-related death (as an indicator of severe liver disease). For the outcome of decompensation, we conducted a Fine-Gray analysis with competing risk of death without decompensation.15 For the endpoint of liver transplant or liver-related death, we conducted a Fine-Gray analysis with liver-related death or liver transplant as the endpoint of interest and non-liver-related death as a competing risk. All models were adjusted for age, sex, disease etiology (non-alcoholic fatty liver disease [NAFLD] or cryptogenic cirrhosis vs. all others, because NAFLD/cryptogenic disease was the most common), and genetic principal components 1-10 to account for ethnic differences. We conducted several sensitivity analyses in which we adjusted for severity of baseline liver disease based on albumin level, platelet count, MELD score, and platelet count and MELD score. We also generated models stratified by presence of clinically significant portal hypertension (CSPH), defined by either liver stiffness measurement ≥25 kPa on VCTE or presence of varices or collaterals on endoscopy or imaging.17 Pre-specified subgroup analyses were: sex (female vs. male), etiology (NAFLD/cryptogenic vs. other), age (≥ or <60 years), presence of diabetes, and presence of severe obesity (class 2-3) vs. not. Heterogeneity between the subgroups was computed based on the Cochran Q statistic.16
Analyses were conducted using R version 4.0.2 (Vienna, Austria) with competing risk analyses conducted using the cmprsk package. A 2-sided p value <0.05 was used to determine statistical significance throughout.
Results
Cohort
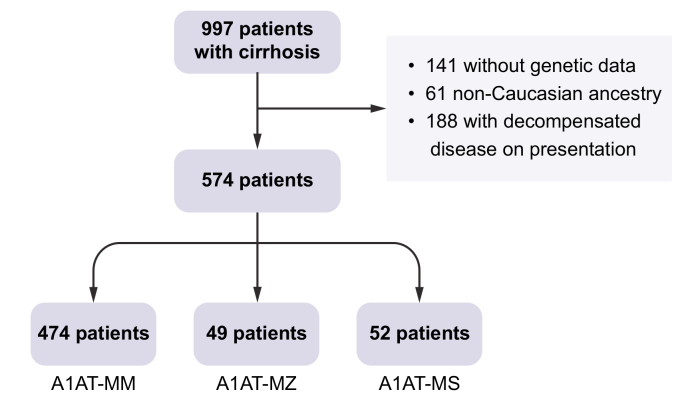
This is a retrospective analysis of patients enrolled in MGI. We included 574 patients with baseline compensated cirrhosis (i.e. who had no evidence of decompensation until at least 6 months after cirrhosis diagnosis) who had undergone genotyping (Fig. 1). We also included only Caucasian participants to avoid attributing effects to these genotypes that may be related to race, as the most common A1AT deficiency-associated variants, Pi∗Z and Pi∗S, are rare in non-Caucasians. Of these 574 patients, 474 had Pi∗MM, 49 had Pi∗MZ, and 52 had Pi∗MS genotypes. In subsequent analyses, we excluded patients with Pi∗ZZ, Pi∗SZ, and Pi∗SS genotypes due to the small number of these patients (n = 9 combined). Of the remaining participants (i.e. with Pi∗MM, Pi∗MZ, or Pi∗MS genotypes), 278 eventually developed hepatic decompensation, 165 died, and 98 underwent liver transplantation >6 months from enrollment. First decompensating event was ascites in 107 patients, hepatic encephalopathy in 101, variceal bleeding in 23, and multiple decompensations in the remainder. Cause of death was liver-related in 51% of deaths, cardiovascular in 8%, and unknown/other in the remainder. Median follow-up was 58.4 months (interquartile range 29.4 to 99.5 months). Incidence rate for decompensation was 0.11 per person-year at risk.
Fig. 1.
Study design.
A1AT, alpha-1 antitrypsin.
Baseline characteristics
Baseline characteristics based on A1AT genotype are shown in Table 1. In the overall cohort, median age was 58 years and 42% of patients were female. The most common etiology of liver disease was NAFLD (45%), followed by alcohol-related liver disease (18%), hepatitis C virus (14%), and hepatitis C combined with alcohol (9%). Laboratory values were as expected for a cohort of baseline compensated cirrhosis, with low to borderline-low platelet count and low MELD scores. There were no meaningful differences in laboratory values or comorbidities across the different A1AT genotypes (Table 1).
Table 1.
Baseline characteristics based on alpha-1 antitrypsin genotype.
| Trait | Pi∗MM, n = 474 | Pi∗MZ, n = 49 | Pi∗MS, n = 52 | p value |
|---|---|---|---|---|
| Age (years) | 58.1 (51.1-65.0) | 59.5 (43.9-66.2) | 57.8 (50.1-67.0) | 0.99 |
| Male | 59.5% | 49.0% | 52.9% | 0.20 |
| Hypertension | 70.3% | 65.3% | 80.4% | 0.58 |
| Hyperlipidemia | 47.9% | 53.1% | 56.9% | 0.59 |
| Diabetes | 53.6% | 63.3% | 47.1% | 0.25 |
| Coronary artery disease | 25.3% | 25.0% | 22.0% | 1.0 |
| Cerebrovascular disease | 6.2% | 4.2% | 6.0% | 0.80 |
| Body mass index (kg/m2) | 31.4 (26.4-36.9) | 30.7 (27.3-38.5) | 31.6 (27.5-36.1) | 0.69 |
| Smoking history | ||||
| Never | 41.4% | 44.9% | 37.3% | 0.31 |
| Former | 36.3% | 44.9% | 39.2% | |
| Current | 22.4% | 10.2% | 23.5% | |
| Illicit drug use history | 14.5% | 8.3% | 3.9% | 0.34 |
| Cirrhosis etiology | ||||
| Non-alcoholic fatty liver disease or cryptogenic | 44.1% | 40.8% | 54.9% | 0.15 |
| Hepatitis C alone | 15.3% | 10.4% | 13.7% | |
| Alcohol alone | 18.1% | 16.3% | 15.7% | |
| Hepatitis C and alcohol | 9.4% | 4.2% | 9.8% | |
| Other | 13.7% | 28.6% | 5.9% | |
| Gastroesophageal varices | 47.0% | 42.6% | 46.0% | 0.67 |
| Clinically significant portal hypertension |
48.5% |
46.8% |
46.0% |
0.92 |
| Laboratory values | ||||
| Creatinine (mg/dl) | 0.9 (0.7-1.1) | 0.8 (0.7-1.0) | 0.9 (0.8-1.1) | 0.36 |
| Hemoglobin A1c (%) | 6.1 (5.4-7.8) | 7.1 (5.9-7.8) | 5.8 (5.2-7.2) | 0.13 |
| Alanine aminotransferase (U/L) | 43.0 (26.0-73.0) | 39.0 (28.0-57.0) | 35.0 (23.0-51.5) | 0.10 |
| Aspartate aminotransferase (U/L) | 49.0 (33.0-79.5) | 54.0 (41.0-71.0) | 46.0 (28.0-78.5) | 0.48 |
| Total bilirubin (mg/dl) | 0.8 (0.5-1.5) | 1.1 (0.7-2.0) | 0.8 (0.5-1.5) | 0.037 |
| Alkaline phosphatase (U/L) | 119.0 (85.0-172.5) | 124.0 (90.0-166.0) | 109.0 (81.5-138.5) | 0.39 |
| Albumin (mg/dl) | 3.9 (3.4-4.3) | 3.9 (2.9-4.2) | 4.0 (3.5-4.3) | 0.76 |
| Platelets (K/ul) | 129.0 (97.0-194.8) | 140.0 (105.0-198.0) | 161.0 (97.5-238.0) | 0.17 |
| Model for end-stage liver disease score | 8.5 (7.2-11.9) | 10.0 (7.7-13.1) | 8.2 (7.2-13.8) | 0.29 |
Three-way comparisons of continuous variables were performed with Kruskal-Wallis statistics, and comparisons of categorical variables were performed using a chi-squared statistic.
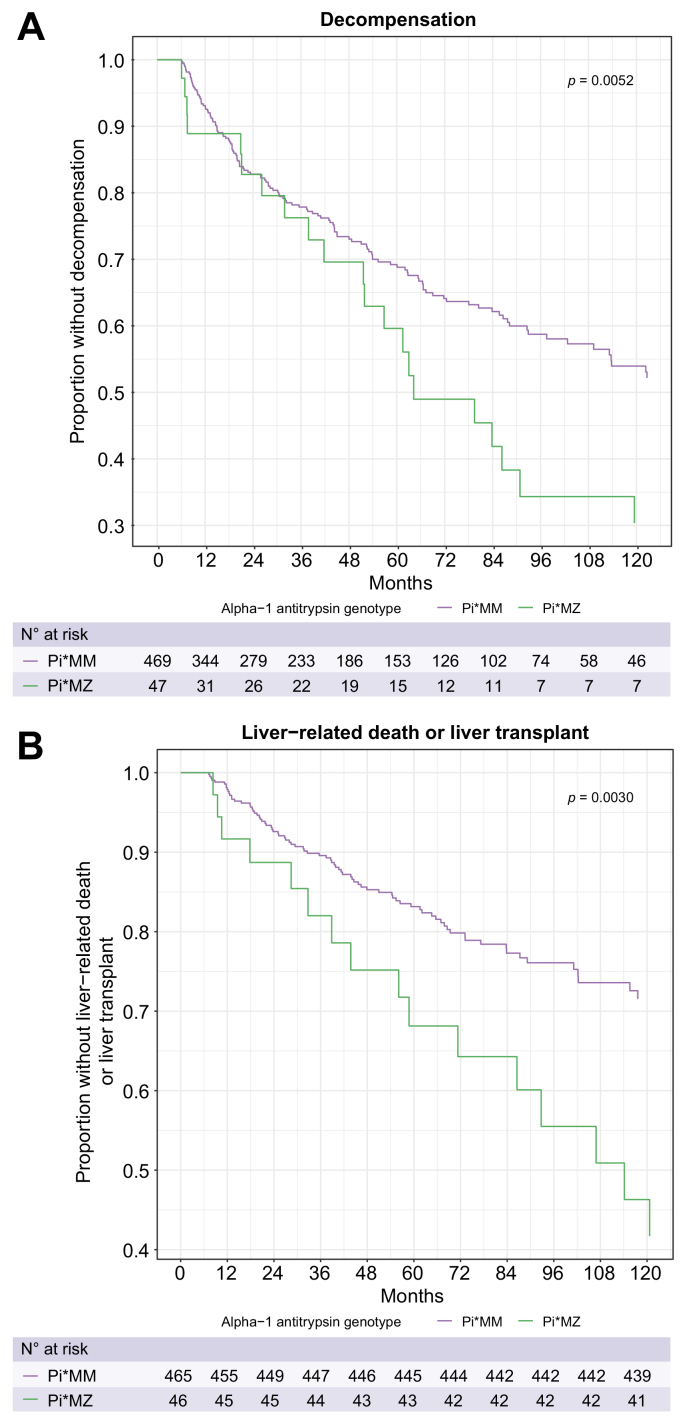
Effects of A1AT genotype on hepatic decompensation and liver-related death or transplant
Compared to Pi∗MM, the Pi∗MZ genotype was associated with increased risk of hepatic decompensation in the primary model with hazard ratio (HR) 1.81 (95% CI 1.22-2.69, p = 0.0032) (Table 2, Fig. 2). This effect was not meaningfully changed after adjusting for severity of underlying liver disease based on albumin, platelet count, and/or MELD score (Table 2). In addition, Pi∗MZ genotype was associated with an increased risk for liver transplantation or liver-related death with HR 2.07 (95% CI 1.21-3.52, p = 0.0078) relative to Pi∗MM (Table 2). These associations remained significant after adjustment for albumin concentration, platelet count, MELD score, or platelet count and MELD score. In contrast, Pi∗MS was not significantly associated with hepatic decompensation or the combined outcome of liver transplant or liver-related death relative to Pi∗MM (Table S2, Fig. S1.).
Table 2.
Effects of Pi∗MZ genotype on hepatic decompensation and liver transplant or liver-related death.
| Model | Hepatic decompensation |
Liver transplant or liver-related death |
||
|---|---|---|---|---|
| Hazard ratio | p value | Hazard ratio | p value | |
| Model 1 | 1.81 (1.22-2.69) | 0.0032 | 2.07 (1.21-3.52) | 0.0078 |
| Model 2 | 1.80 (1.22-2.65) | 0.0030 | 1.86 (1.07-3.23) | 0.027 |
| Model 3 | 1.80 (1.22-2.65) | 0.003 | 1.92 (1.10-3.36) | 0.021 |
| Model 4 | 1.79 (1.19-2.67) | 0.0047 | 1.90 (1.10-3.28) | 0.022 |
| Model 5 | 1.80 (1.22-2.68) | 0.0033 | 1.80 (1.02-3.18) | 0.041 |
Hazard ratios (95% Cl) for Pi∗MZ relative to Pi∗MM. Model 1 is adjusted for age, sex, disease etiology (non-alcoholic fatty liver disease or cryptogenic vs. other etiologies), and principal components 1-10. Model 2 is adjusted for model 1 covariates plus albumin. Model 3 is adjusted for model 1 covariates plus platelet count. Model 4 is adjusted for model 1 covariates plus model for end-stage liver disease score. Model 5 is adjusted for model 1 covariates plus platelet count and model for end-stage liver disease score. p value was generated using a Fine-Gray competing risk model.
Fig. 2.
Effects of SERPINA1 Pi∗MZ genotype on liver-related outcomes. (A) Fine-Gray competing risk model depicting time to hepatic decompensation, with competing risk of death without hepatic decompensation. (B) Fine-Gray competing risk model depicting time to liver-related death or liver transplant, with competing risk of non-liver-related death. p values are by the Fine-Gray method.
We evaluated the association between the Pi∗MZ genotype and decompensation or liver-related death or transplant in several subgroups (Table S3). As expected, most of the effects were less significant in subgroups compared to the overall cohort due to smaller sample size, but all odds ratios were directionally consistent. Pi∗MZ was significantly associated with hepatic decompensation in patients with but not without baseline CSPH. We note that the association between Pi∗MZ and decompensation has greater magnitude in men compared to women, and may also be greater in older vs. younger individuals, though heterogeneity was not statistically significant for any comparison (phet >0.05). In addition, the association between Pi∗MZ and decompensation or liver-related death/transplant was similar across disease etiologies. Alcohol intake at time of cirrhosis diagnosis was quantified in only 29% of patients; in a model adjusting for alcohol intake, Pi∗MZ was not significantly associated with decompensation (p >0.05 for all associations).
Effects of A1AT genotype on individual decompensating events
We evaluated the impact of Pi∗MZ on the individual decompensating events, namely ascites, hepatic encephalopathy, and variceal bleeding (Table S4). The association was significant for ascites (HR 1.82, 95% CI 1.09-3.05, p = 0.022) and trending toward significance for hepatic encephalopathy (HR 1.54, 95% CI 0.97-2.43, p = 0.065), and was directionally consistent for variceal bleeding though there were few incident events (n = 40 with incident events >6 months after follow-up).
Discussion
We found that compared to A1AT Pi∗MM, the Pi∗MZ genotype was associated with increased hepatic decompensation and increased risk of severe liver disease (defined as liver-related death or liver transplant) in patients with baseline cirrhosis. This effect was robust across a number of subgroups and for individual outcomes. In contrast, Pi∗MS was not associated with decompensation or death/liver transplant. To our knowledge, this is the first study showing that in patients with compensated cirrhosis, Pi∗MZ is associated with an increased rate of progression to decompensation or the composite outcome of death/liver transplantation.
The association between Pi∗Z and development of cirrhosis and/or decompensation in the general population is well-established, but there are few data on decompensation among those with established cirrhosis. We are only aware of one other study evaluating effects of Pi∗MZ on decompensation focusing on those with cirrhosis. That study included 540 individuals with Pi∗MM or Pi∗MZ genotypes and found that Pi∗MZ carriers had increased odds of ascites and hepatic encephalopathy and a higher MELD score, with no overall difference in transplant-free survival.10 It differed from our study in that it was cross-sectional and therefore was unable to evaluate whether Pi∗MZ patients were simply presenting at a later timepoint in their disease. Our study, in contrast, identified patients at a similar disease stage, all compensated at baseline, and found that Pi∗MZ was associated with increased risk of incident outcomes. Therefore, our study offers information in terms of disease trajectory even for patients at a similar initial disease stage.
The clinical implications of this study are that A1AT Pi∗MZ can be treated as a second risk factor for disease progression. In our cohort, 8% of Caucasian patients with cirrhosis had the Pi∗MZ risk allele. Notably, the Pi∗Z allele frequency is higher in those with cirrhosis than in the general population because Pi∗MZ is also associated with increased risk of cirrhosis. It is worth comparing the effects of the Pi∗MZ genotype with that of diabetes, the metabolic comorbidity most strongly linked to cirrhosis and hepatic decompensation.18 Among the few studies on diabetes or impaired glucose tolerance as a risk factor for decompensation in compensated cirrhosis, the HR for decompensation was 1.4-1.7,19,20 which is comparable in magnitude to the observed effect of A1AT Pi∗MZ. Of note, the Pi∗MZ allele is known to interact with other risk factors such as diabetes and obesity to increase the risk of incident cirrhosis,9 implying that treatment of these modifiable comorbidities may mitigate the non-modifiable genetic risk factor of Pi∗MZ. While missing data prevented us from assessing the interaction of alcohol consumption and Pi∗MZ, strict alcohol abstinence may be even more important in Pi∗MZ patients with cirrhosis. Whether treatment of other liver diseases such as viral hepatitis or obesity decreases risk of decompensation in Pi∗MZ carriers with established cirrhosis more than in Pi∗MM carriers is unknown but plausible.
Limitations of this study include that it was a single center study. In addition, even with manual chart review, it is difficult to know precisely when decompensation developed in a retrospective study. The prevalence of diabetes and obesity was high in this cohort, suggesting that many of our patients may have had underlying undiagnosed NAFLD, and this may limit generalizability to populations with lower levels of metabolic comorbidities. We included only Caucasian patients because the Pi∗Z and Pi∗S alleles are very rare in other races and we wanted to avoid confounding our findings with other effects of race including social determinants of health. While this exclusion limits generalizability to other races, the Pi∗Z and Pi∗S alleles are very rare in non-Caucasian populations, so at a population level they are unlikely to be as important. Alcohol intake from time of cirrhosis diagnosis was not quantified in a large proportion of patients, which limited our ability to determine how Pi∗MZ may interact with alcohol intake. Decompensation rates were higher than would be expected, likely due to the high prevalence of diabetes and obesity and the retrospective nature of this study which may have included a higher number of patients who were followed for longer and therefore had more events. However, the design of MGI did not select for patients with advanced liver disease or those with more frequent encounters with our hospital, as most of the patients in this study were recruited at the time of elective surgery. Finally, since this was a retrospective study, it is possible that some patients had decompensation at baseline but were not coded as such. However, the miscoding would be identified by our chart review and would also occur in patients with Pi∗MM and Pi∗MS, not just those with Pi∗MZ. Furthermore, most decompensating events occurred more than 3 years after the initial diagnosis of cirrhosis. Strengths of our study include detailed phenotyping with manual chart review and robust statistical analyses including a 6-month lead time for outcomes and adjustment for relevant covariables including ethnicity via principal components.
In conclusion, the A1AT Pi∗MZ genotype is associated with increased risk of hepatic decompensation and transplant/liver-related death in a cohort of patients with compensated cirrhosis at baseline. Further multicenter studies to validate these findings and determine whether more aggressive intervention can decrease the risk of adverse outcomes in Pi∗MZ carriers are warranted.
Financial support
This study was supported in part by a Clinical, Translational and Outcomes Research Award from the American Association for the Study of Liver Diseases to VLC.
Authors’ contributions
Vincent L. Chen: concept development, study design, data analysis, drafting of manuscript. Daniel A. Burkholder: data collection, data analysis. Isabel J. Moran: data collection, data analysis. Jacob V. DiBattista: data collection, critical review of the manuscript. Matthew S. Miller: data collection, critical review of the manuscript. Yanhua Chen: data analysis, critical review of the manuscript. Xiaomeng Du: data analysis, critical review of the manuscript. Anna Lok: critical review of the manuscript. Elizabeth Speliotes: concept development, study design, critical review of the manuscript. All authors approved the final version of the article, including the authorship list.
Data availability statement
Individual-level data will not be made available due to patient privacy restrictions. Summary data will be made available upon reasonable request to the corresponding author.
Conflicts of interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100483.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Greene C.M., Marciniak S.J., Teckman J., Ferrarotti I., Brantly M.L., Lomas D.A., et al. alpha1-Antitrypsin deficiency. Nat Rev Dis Primers. 2016;2:16051. doi: 10.1038/nrdp.2016.51. [DOI] [PubMed] [Google Scholar]
- 2.Stoller J.K., Brantly M. The challenge of detecting alpha-1 antitrypsin deficiency. COPD. 2013;10(Suppl 1):26–34. doi: 10.3109/15412555.2013.763782. [DOI] [PubMed] [Google Scholar]
- 3.Piitulainen E., Tanash H.A. The clinical profile of subjects included in the Swedish National Register on individuals with severe alpha 1-antitrypsin deficiency. COPD. 2015;12(Suppl 1):36–41. doi: 10.3109/15412555.2015.1021909. [DOI] [PubMed] [Google Scholar]
- 4.Perlmutter D.H., Brodsky J.L., Balistreri W.F., Trapnell B.C. Molecular pathogenesis of alpha-1-antitrypsin deficiency-associated liver disease: a meeting review. Hepatology. 2007;45:1313–1323. doi: 10.1002/hep.21628. [DOI] [PubMed] [Google Scholar]
- 5.Chen V.L., Chen Y., Du X., Handelman S.K., Speliotes E.K. Genetic variants that associate with cirrhosis have pleiotropic effects on human traits. Liver Int. 2019;40:405–415. doi: 10.1111/liv.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark V.C., Marek G., Liu C., Collinsworth A., Shuster J., Kurtz T., et al. Clinical and histologic features of adults with alpha-1 antitrypsin deficiency in a non-cirrhotic cohort. J Hepatol. 2018;69:1357–1364. doi: 10.1016/j.jhep.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Tanash H.A., Piitulainen E. Liver disease in adults with severe alpha-1-antitrypsin deficiency. J Gastroenterol. 2019;54:541–548. doi: 10.1007/s00535-019-01548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamesch K., Mandorfer M., Pereira V.M., Moeller L.S., Pons M., Dolman G.E., et al. Liver fibrosis and metabolic alterations in adults with alpha-1-antitrypsin deficiency caused by the Pi∗ZZ mutation. Gastroenterology. 2019;157:705–719 e718. doi: 10.1053/j.gastro.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Fromme M., Schneider C.V., Pereira V., Hamesch K., Pons M., Reichert M.C., et al. Hepatobiliary phenotypes of adults with alpha-1 antitrypsin deficiency. Gut. 2021 doi: 10.1136/gutjnl-2020-323729. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer B., Mandorfer M., Viveiros A., Finkenstedt A., Ferenci P., Schneeberger S., et al. Heterozygosity for the alpha-1-antitrypsin Z allele in cirrhosis is associated with more advanced disease. Liver Transpl. 2018;24:744–751. doi: 10.1002/lt.25057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey R., Schmidt E.M., Abecasis G.R., Lee S. A fast and accurate algorithm to test for binary phenotypes and its application to PheWAS. Am J Hum Genet. 2017;101:37–49. doi: 10.1016/j.ajhg.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen V.L., Du X., Chen Y., Kuppa A., Handelman S.K., Vohnoutka R.B., et al. Genome-wide association study of serum liver enzymes implicates diverse metabolic and liver pathology. Nat Commun. 2021;12:816. doi: 10.1038/s41467-020-20870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu C., Caussy C., Imajo K., Chen J., Singh S., Kaulback K., et al. Magnetic resonance vs transient elastography analysis of patients with nonalcoholic fatty liver disease: a systematic review and pooled analysis of individual participants. Clin Gastroenterol Hepatol. 2019;17:630–637 e638. doi: 10.1016/j.cgh.2018.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran I.J., Burkholder D.A., DiBattista J.V., Chen V.L. 92 Specificity and positive predictive value of International Classification of Diseases-10 diagnosis codes for cirrhosis. Gastroenterology. 2021;160 S-762-S-763. [Google Scholar]
- 15.Fine J.P., Gray R.J. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 16.Cochran W.G. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 17.de Franchis R., Bosch J., Garcia-Tsao G., Reiberger T., Ripoll C., Abraldes J.G., et al. Baveno VII - renewing consensus in portal hypertension. J Hepatol. 2021 doi: 10.1016/j.jhep.2021.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis H., Craig D., Barker R., Spiers G., Stow D., Anstee Q.M., et al. Metabolic risk factors and incident advanced liver disease in non-alcoholic fatty liver disease (NAFLD): a systematic review and meta-analysis of population-based observational studies. Plos Med. 2020;17 doi: 10.1371/journal.pmed.1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calzadilla-Bertot L., Vilar-Gomez E., Torres-Gonzalez A., Socias-Lopez M., Diago M., Adams L.A., et al. Impaired glucose metabolism increases risk of hepatic decompensation and death in patients with compensated hepatitis C virus-related cirrhosis. Dig Liver Dis. 2016;48:283–290. doi: 10.1016/j.dld.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Saeed M.J., Olsen M.A., Powderly W.G., Presti R.M. Diabetes mellitus is associated with higher risk of developing decompensated cirrhosis in chronic hepatitis C patients. J Clin Gastroenterol. 2017;51:70–76. doi: 10.1097/MCG.0000000000000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual-level data will not be made available due to patient privacy restrictions. Summary data will be made available upon reasonable request to the corresponding author.