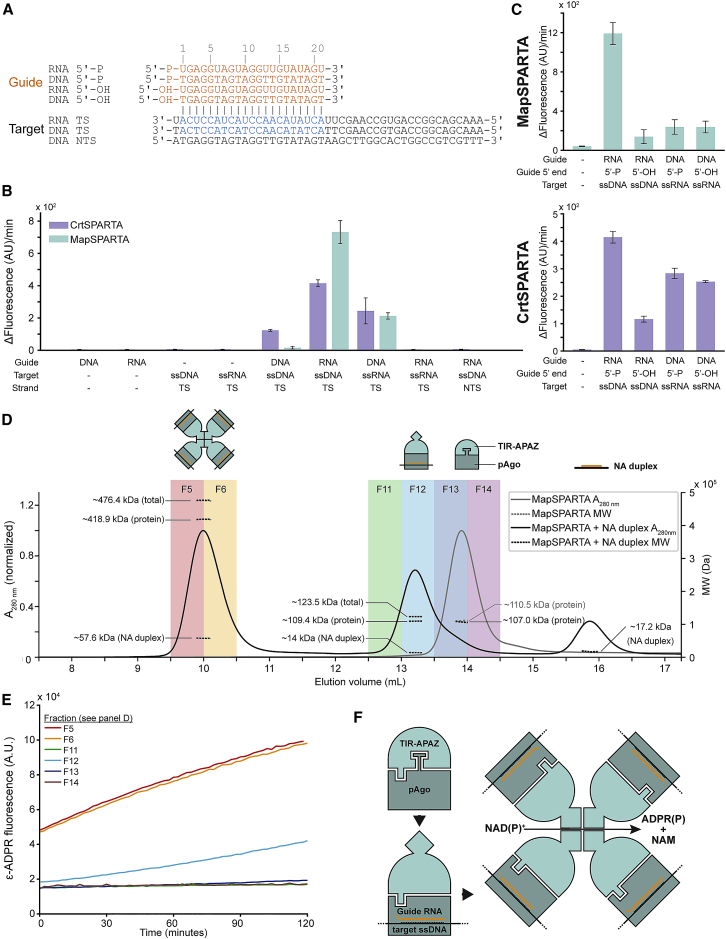
Figure 3.
SPARTA degrades NAD+ upon RNA-guided ssDNA binding
(A) Schematic representation of guide (orange) and target (gray, target sequence: blue) oligonucleotides used in (B) and (C).
(B and C) MapSPARTA or CrtSPARTA was mixed with guide and target oligonucleotides in a 1:1:2 molar ratio and incubated with ϵ-NAD+. Graphs indicate the maximum change in fluorescence over time. The average of three technical replicates is shown; error bars indicate standard deviations.
(B) SPARTA is preferentially activated upon by guide RNA-mediated ssDNA target binding. (C) SPARTA prefers 5′-P guides over 5′-OH guides. CrtSPARTA 5′-P guide samples are identical in (B) and (C).
(D) SPARTA oligomerizes upon guide RNA-mediated target ssDNA binding. MapSPARTA was incubated with guide RNA and target ssDNA in a 1:1:1 molar ratio and subjected to size exclusion chromatography-multi-angle light scattering analysis. Colored backgrounds indicate fractions that were tested for NADase activity (E). Control without nucleic acids is identical to that in Figure 2C.
(E) SPARTA is catalytically activated upon oligomerization. SEC-MALS fractions (D) were incubated with ϵ-NAD+, and change in fluorescence was determined over time.
(F) Proposed mechanism for SPARTA activation. TIR-APAZ and associated short pAgo form a catalytically inactive heterodimeric SPARTA complex. Guide RNA-mediated target ssDNA binding induces conformational changes that facilitate oligomerization. Guide/target-bound SPARTA heterodimers form a complex with a 4:4:4:4 (TIR-APAZ:short pAgo:guide:target) stoichiometry in which the NAD(P)ase activity of the TIR domain is unleashed.
See also Figure S3.