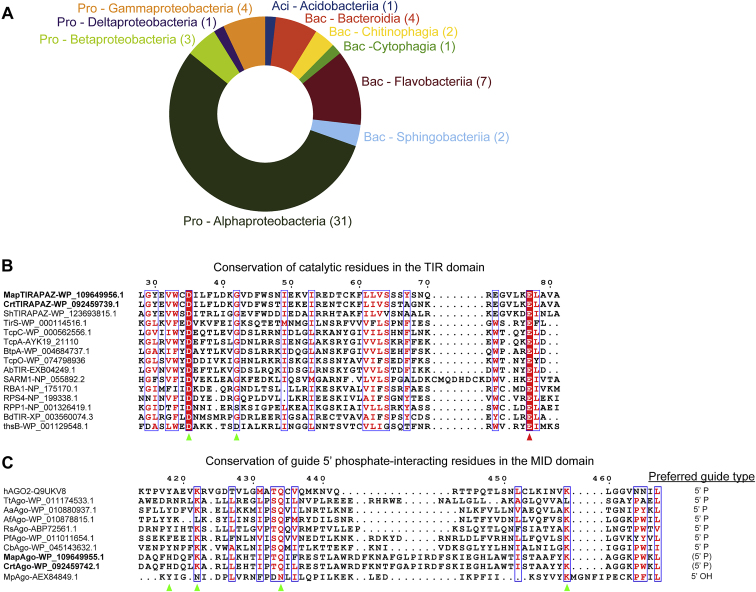
Figure S1.
Distribution and conservation of catalytic residues in SPARTA systems, related to Figure 1
(A) Circle diagram indicating bacterial phyla (abbreviated) and classes in which SPARTA systems are found. Aci, acidobacteria; Bac, bacteroidetes; Pro, proteobacteria.
(B) Multiple sequence alignment of TIR domains from SPARTA systems with various prokaryotic and eukaryotic TIR domains that demonstrate NADase activity. Green and red triangles indicate catalytic residues important for NADase activity; red triangle indicates residue substituted in CrtTIR-APAZE77A and MapTIR-APAZE77A.
(C) Multiple sequence alignment of MID domains from different prokaryotic Argonaute proteins and their preferred guide types. Green triangles indicate residues important for 5′-phosphate binding.