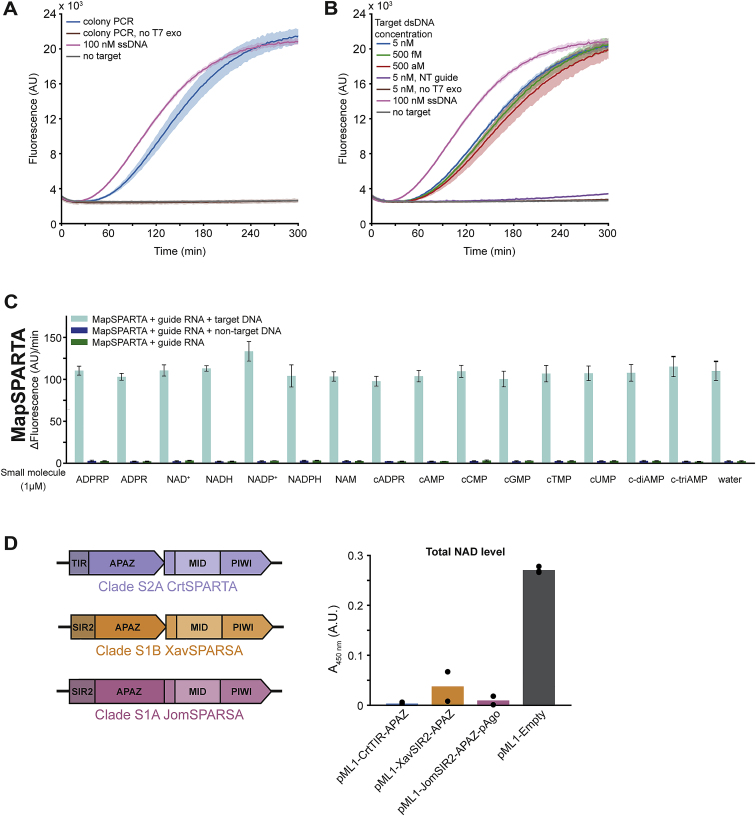
Figure S7.
SPARTA can be used to detect dsDNA, and SPARSA systems degrade NAD+in vivo, related to Figure 7
(A) SPARTA combined with colony PCR and subsequent T7 exonuclease treatment facilitates the detection of dsDNA. MapSPARTA was mixed with guide RNA in a 1:1 molar ratio, and colony PCR-amplified DNA treated with T7 exonuclease was added. After the addition of ε-NAD+, total fluorescence was measured over time. No target negative control and 100 nM ssDNA positive control are identical to those in Figure 7F.
(B) SPARTA mixed with target dsDNA, and T7 exonuclease simultaneously facilitates the detection of dsDNA at aM levels. MapSPARTA was mixed with guide RNA in a 1:1 molar ratio, and target DNA and T7 exonuclease were added. After the addition of ε-NAD+, total fluorescence was measured over time. No target negative control and 100 nM ssDNA positive control are identical to those in Figure 7F.
(C) MapSPARTA is not activated by small molecules, and small molecules do not enhance guide RNA/target ssDNA-mediated activity of MapSPARTA. MapSPARTA was mixed with guide and target oligonucleotides in a 1:1:2 molar ratio and incubated with ϵ-NAD+. Graphs indicate the maximum change in fluorescence over time.
(D) Left: schematic diagram of the operon structure and domain organization of SPARTA, SPARSA-1A, and SPARSA-1B systems. Right: SPARSA SIR2-APAZ proteins lower total NAD when heterologously expressed in E. coli. Total NAD levels were determined in E. coli cultures harboring an empty expression vector (pML1) or a pML1 expression vector encoding clade S2A CrtTIR-APAZ, clade S1A Joostella marina JomSIR2-APAZ-Ago, or clade S1B Xanthomonas vesicatoria XavSIR2-APAZ. Graphs show the average of three (A and C) or two (B) technical replicates or two biological replicates (D). Error bars and shadings indicate standard deviations.