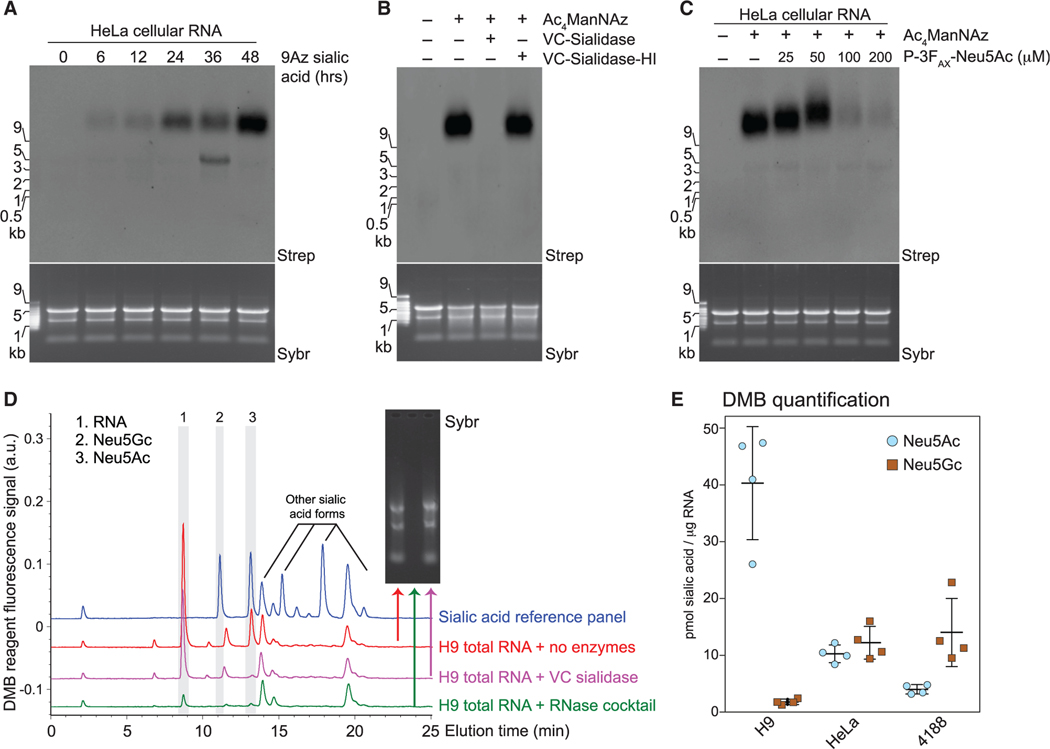
Figure 3. Glycans modifying RNA contain sialic acid.
(A) Blotting of RNA from HeLa cells treated with 1.75 mM 9-azido sialic acid for indicated times.
(B) Blotting of Ac4ManNAz-labeled HeLa cell RNA treated with Vibrio cholerae (VC) sialidase or heat-inactivated sialidase (VC-sialidase-HI).
(C) Blotting of RNA from HeLa cells treated with Ac4ManNAz and the indicated concentrations of P-3FAX-Neu5Ac.
(D) Unlabeled total RNA from H9 cells was isolated, reacted with the indicated enzyme (no enzymes, RNase cocktail, or Sialidase treatment), cleaned up to remove cleaved metabolites, and processed with the fluorogenic 1,2-diamino-4,5-methylenedioxybenzene (DMB) probe. HPLC analysis quantified the presence and abundance of specific sialic acids. Inset, Sybr gel image of the total RNA for each condition. The main sialic acid peaks are 2 and 3. The identity of peak 1 is unknown, but it is RNase-sensitive.
(E) Quantification of DMB results (D) from 4,188, H9, and HeLa cells from four biological replicates.
See also Figure S3.