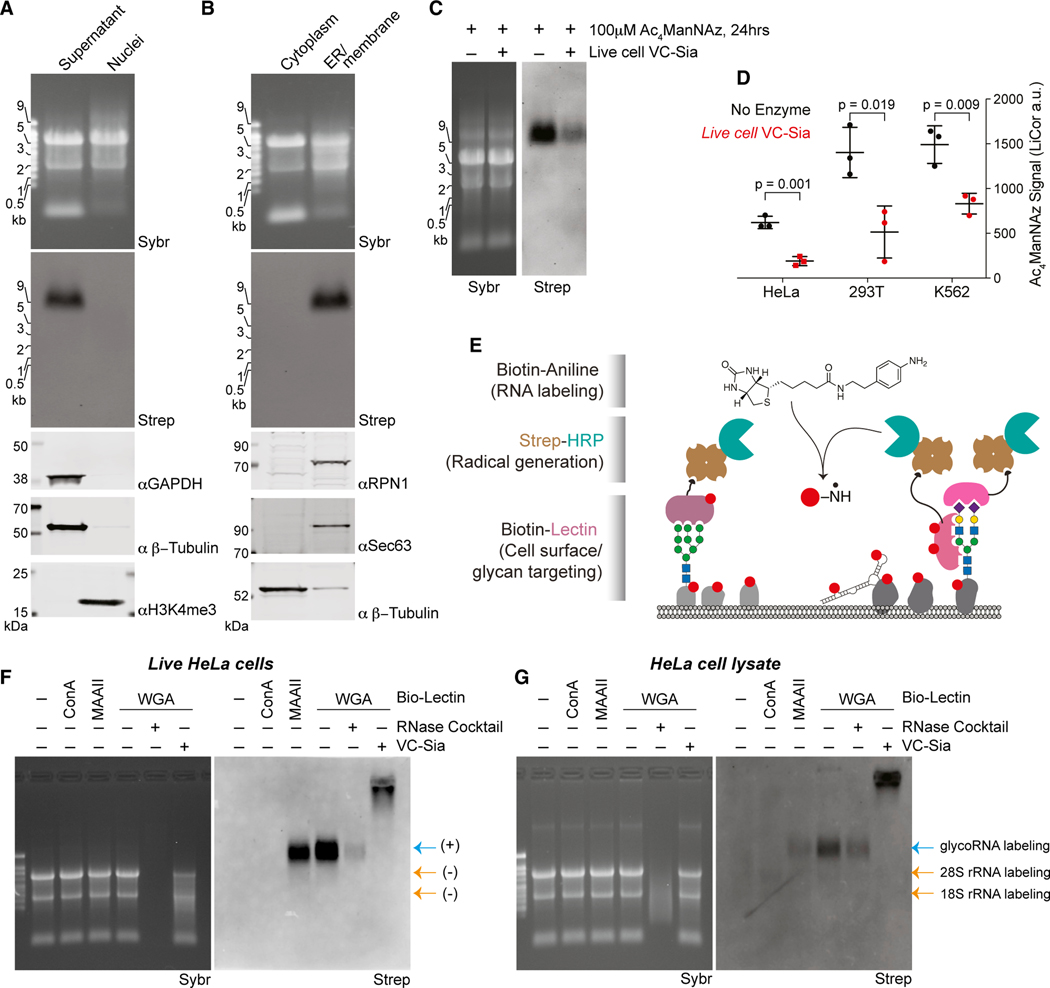
Figure 5. glycoRNAs are on the external surface of living cells.
(A) Blotting of RNA and proteins after subcellular fractionation designed to robustly purify nuclei. Non-nuclear proteins GAPDH and β-tubulin and nuclear histone 3 lysine 4 trimethylation (H3K4me3) are visualized by western blot.
(B) Blotting of RNA and proteins after subcellular fractionation designed to separate soluble cytosol from membranous organelles. Membrane proteins RPN1, Sec63, and soluble β-tubulin are visualized by western blot.
(C) Blotting of RNA from HeLa cells labeled with 100 μM Ac4ManNAz for 24 h and then exposed to fresh media containing 100 μM Ac4ManNAz with or without 150 nM VC-Sia for 60 min at 37°C
(D) Quantification of the experiment in (C) across biological triplicates and from 293T or K562 cells treated in the same manner. p value calculated by a paired, two-tailed t test.
(E) Schematic of the lectin-based proximity labeling of RNA on cell surfaces. Living cells are stained with a biotinylated lectin that recruits streptavidin-HRP that is in turn able to generate nitrene radicals from biotin-aniline after the addition of hydrogen peroxide. RNA from these cells is then extracted and analyzed for biotin labeling that reveals if that RNA was in proximity to the lectin.
(F) Blotting of total RNA samples generated as described in (E). Lanes 5 and 6 were processed in vitro (after purifying RNA) with RNase cocktail or VC-Sia to demonstrate any sensitivity of the biotin-aniline signal to these enzymes.
(G) Blotting of total RNA samples similar to (F) however cells were first lysed in a hypotonic buffer, destroying cellular membranes that are normally impermeable to nitrene radicals. Labeling of rRNA is evident here but not in (F).
See also Figure S5.