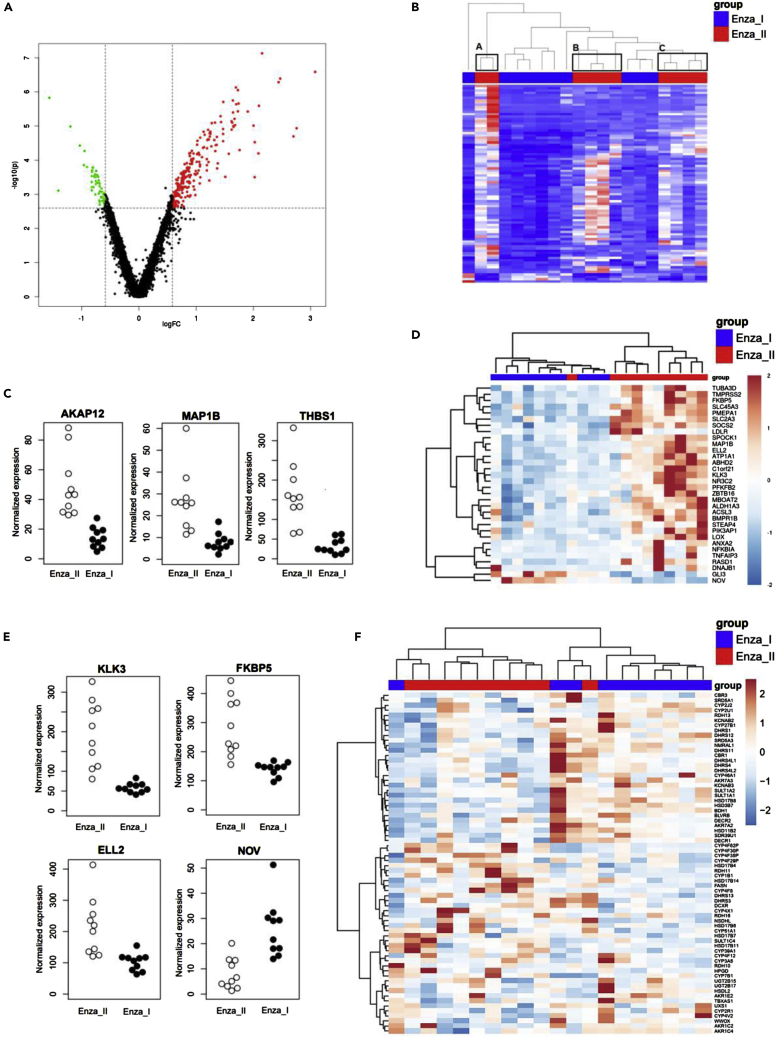
Figure 5.
Enzalutamide resistance is associated with enhanced androgen action as revealed by RNA-seq
The differential gene expression in the tumors between the ORX + Enza I and ORX + Enza II mice by RNA-seq.
(A) By using FC > 1.5 and FDR <0.05, we identified 292 genes with altered expression between the two groups, presented by a volcano plot. Thresholds used in filtering are marked in the plot with dashed lines, upregulated genes (230 genes) are colored red, and downregulated genes (62 genes) are colored green.
(B) Heatmap of hierarchical clustering of the differentially expressed genes in the tumors of the ORX + Enza I and ORX + Enza II mice define three separate clusters for the Enza II mice (A–C). Each row represents one differentially expressed gene, and each column represents one sample.
(C) RNA-seq data of AKAP12, MAP1B and THBS1 representing transcripts with those constantly altered between ORX + Enza I and ORX + Enza II tumors.
(D) Hierarchical clustering of the set of AR-associated genes provides a clear separation of the ORX + Enza I and ORX + Enza II tumors, indicating altered androgen action during the progression of Enza resistance. The heatmaps are based on the differentially expressed AR-related genes (FDR <0.05 and FC >1.5).
(E) RNA-seq data of well-characterized AR-regulated genes (KLK3, FKBP5, ELL2 and NOV) in ORX + Enza I and ORX + Enza II tumors further support the enhanced androgen action in Enza-resistant tumors compared to those with Enza response.
(F) Hierarchical clustering of the RNA-seq data of Enza responsive (ORX + Enza I) and Enza-resistant (ORX + Enza II) tumors using the leading edge genes among enzymes of the CYP, SDR and AKR families potentially involved in steroid metabolism. The leading edge determined which 68 subsets of genes contributed the most to the enrichment signal of a given set of 124 genes. The color gradients (B, D, and F) represents the intensity of gene expression in tumors, with blue indicating low expression in tumor and red indicating high gene expression during ORX + Enza I and ORX+Enza II treatments.