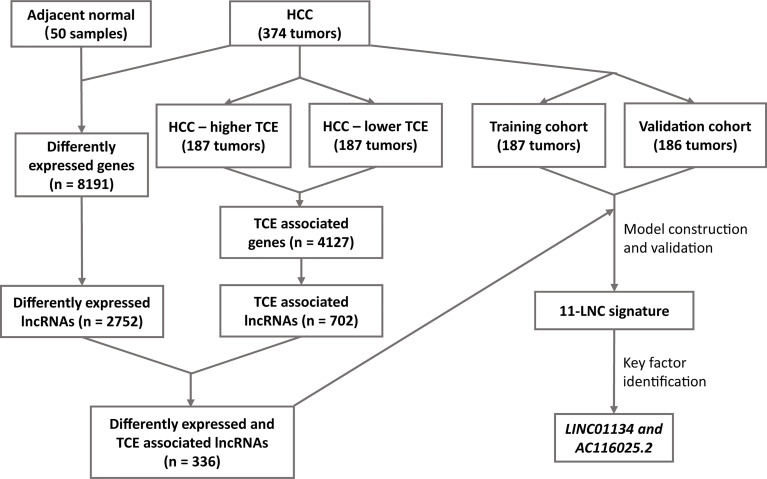
Figure 1.
The workflow of the study. Expression data of hepatocellular carcinoma (HCC) and adjacent normal liver tissues were compared to identify differentially expressed lncRNAs in HCC. All HCC with expression data were divided into higher and lower T cell exclusion (TCE) levels using the TIDE analysis. Higher- and lower-TCE groups were compared to identify TCE-associated lncRNAs. Differentially expressed and TCE-associated lncRNAs were then merged to identify differently expressed and TCE-associated lncRNAs, which were then subjected to LASSO and multivariate Cox analyses to construct the 11LNCPS predictive of patient survival. LINC01134 and AC116025.2 were then identified as the critical members of the signature.