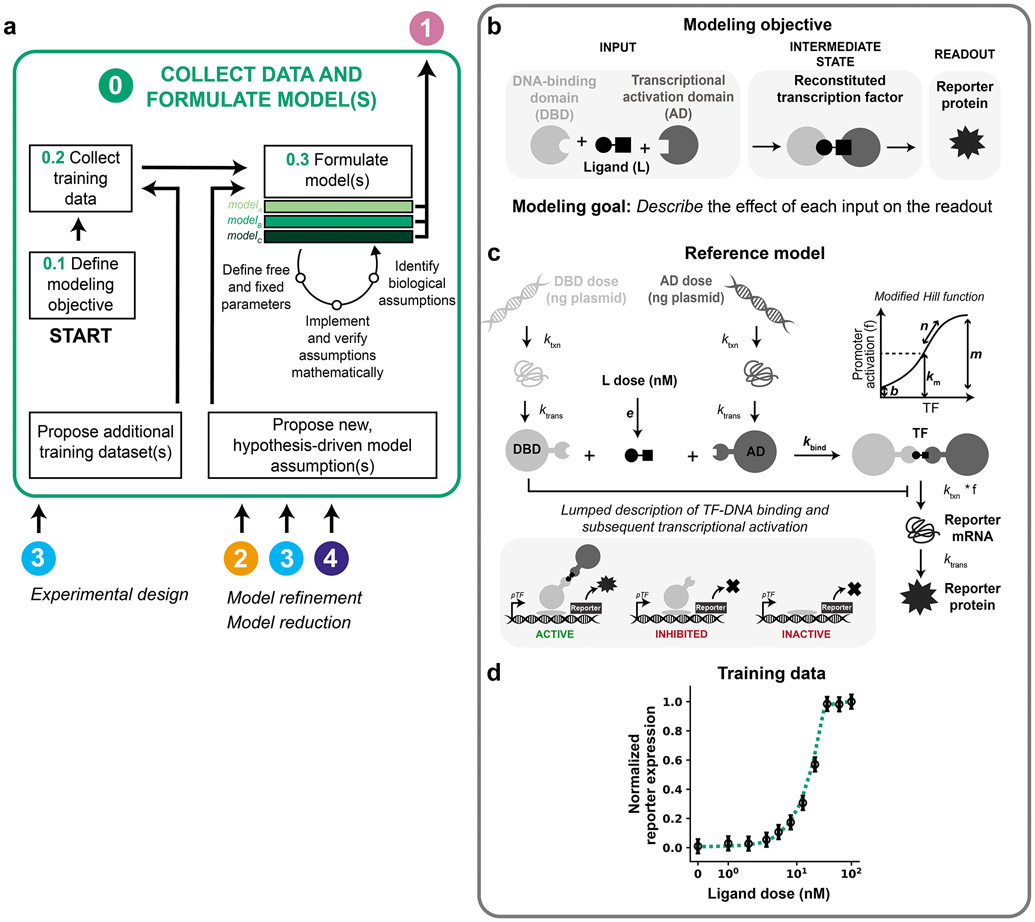
Figure 2. Collect data and formulate model(s).
(a–d) This figure and subsequent figures present the overall workflow (a) and then examine its application to the case study (b–d). (a) Module 0 workflow for collecting training data and formulating model(s). (b–d) Module 0 case study for a hypothetical crTF. (b) Case study modeling objective. The crTF has a DNA-binding domain and an activation domain that reconstitute in the presence of a ligand. The DNA-binding domain and activation domain are each fused to a heterodimerization domain. The reconstituted crTF induces transcription of a reporter gene, and the mRNA translated into a protein that is measured. (c) Schematic of the reference model for the case study. Free parameters are in bold and other parameters are fixed based on literature values (Supplementary Information). Promoter activation is described by a modified Hill function that accounts for these promoter states: active (bound by crTF), inhibited (bound by DNA-binding domain), and inactive (unbound). Degradation reactions are included for all states and for simplicity are not depicted here. Reference parameter values are in Supplementary Table 2. (d) Case study training data. The initial set of training data is a ligand dose response at a constant dose of DNA-binding domain plasmid and activation domain plasmid (50 ng of each plasmid per well of cells). Training data and reference model data are normalized to their respective maximum values. Data points and error bars are the simulated training data with added noise. The dotted line is the reference model. Data are plotted on symmetrical logarithmic (“symlog”) axes in this figure and all subsequent figures for ligand dose response simulations.