Figure 2. Network-based in silico drug repurposing for Alzheimer’s disease (AD).
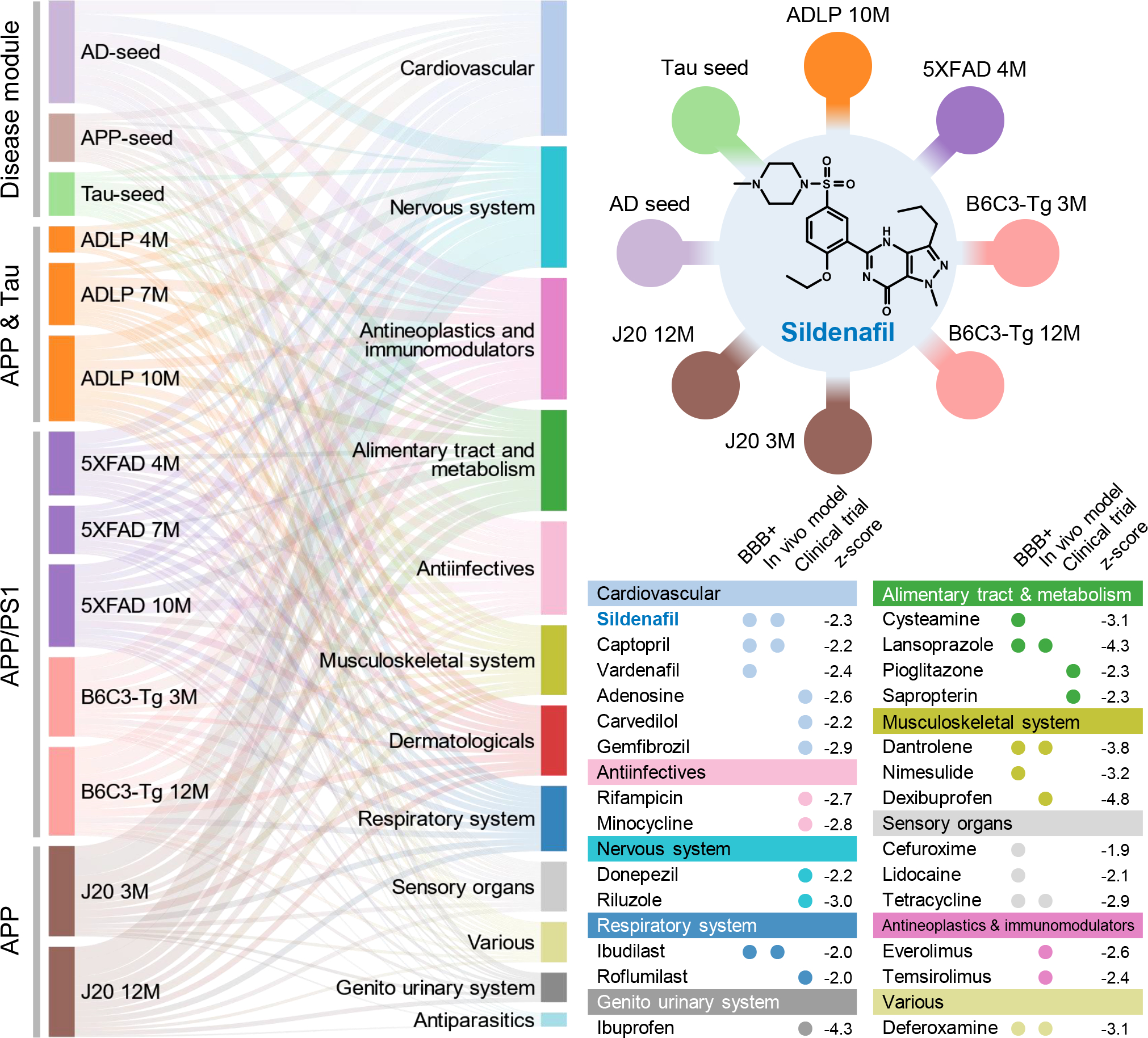
In total, 13 endophenotype disease modules, built by 3 experimentally validated (seed) gene sets in amyloidosis (Amyloid), tauopathy (Tau), and AD, as well as 10 differentially expressed protein from proteomics data generated in AD genetic mouse models, were evaluated to screen FDA-approved drugs using a network proximity measure (Methods). A Sankey diagram illustrates a global view of 66 high-confidence drug candidates, identified by network proximity analysis. Network proximity analysis measures the interplay between endophenotype disease modules (proteins) and drug targets in the human interactome. Drugs are grouped by their first-level Anatomical Therapeutic Chemical Classification (ATC) codes. The drugs with known anti-AD clinical trial status, in vivo animal model, and blood–brain barrier (BBB) properties data are highlighted. The z-score with AD seed module by network proximity is given for each drug. Subject matter expertise criterion-based prioritization resulted in sildenafil as the best candidate (z = −2.30), which has significant close network distance with eight endophenotype disease modules.
