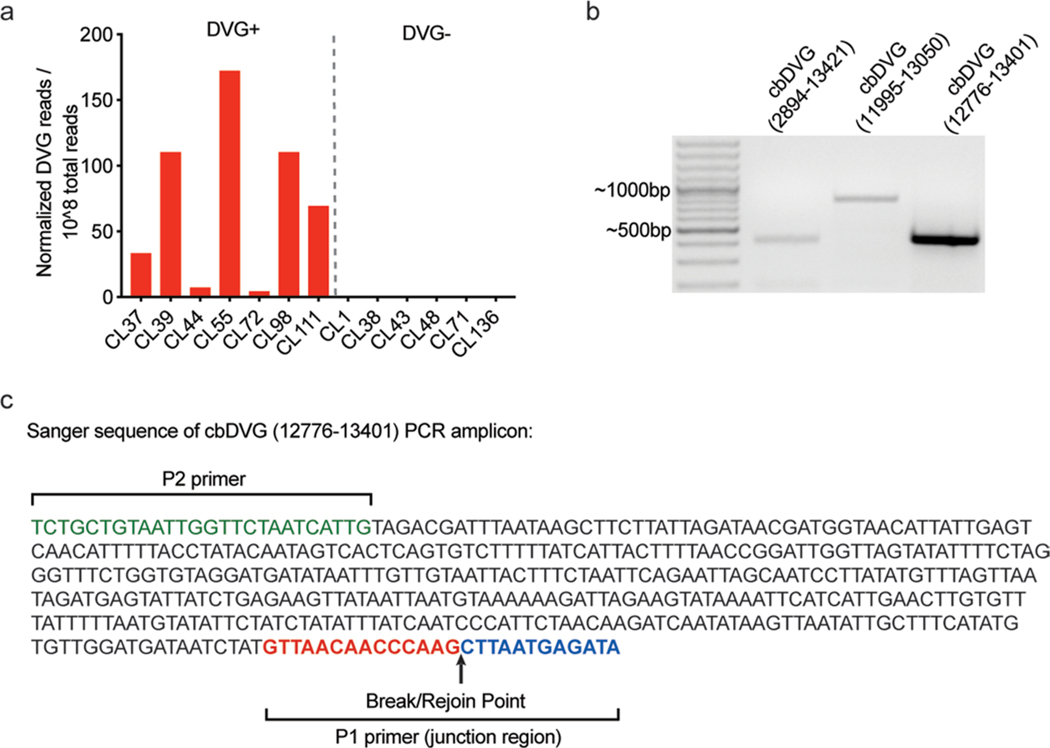
Extended Data Fig. 1 |. Confirmation of rT-PCr and rNA-seq/VoDKA methods with paediatric patient samples.
a, Thirteen hospitalized paediatric patients from cohort 1 were divided into 2 groups based of PCR screening: DVG PCR+ (left) and DVG PCR- (right). DVG reads identified for each patient using the RNA-seq/VODKA pipeline were normalized to 108 total reads. b, Specific primers for unique cbDVG junction regions were designed based on cbDVG sequences identified by RNA-seq/VODKA in sample H75. The numbers in parenthesis indicate break and rejoin positions targeted by the primers in sample H75. Amplicons were of the expected sizes (left to right: 417bp, 804bp and 436bp). c, cbDVG band from sample H75 12776–13401 was gel extracted and confirmed by Sanger’s sequencing. P2 primer sequence is labeled in green and P1 primer sequence (spanning the junction region) is labeled in red and blue to show break/rejoin point.