Figure 1.
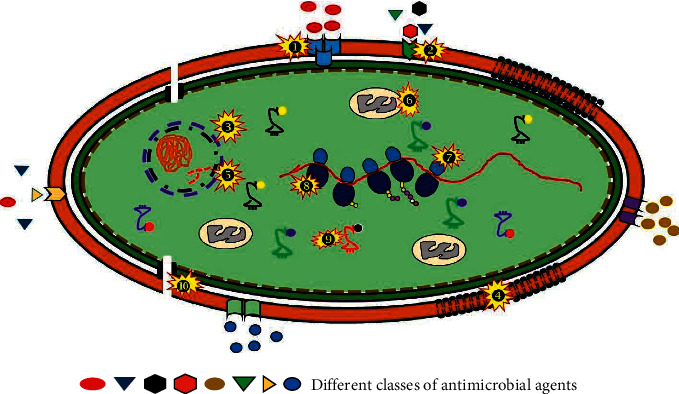
Diagrammatic representation of different target sites for the antimicrobial agent's activity on a bacterial cell. ❶ Alteration in PBP: modified PBP is responsible for the mechanism of action of resistance seen in Gram-positive bacteria as it produces β-lactamases. For example, reduction in affinity of β-lactam antibiotics is due to the mutation on penicillin-binding protein. ❷ Efflux pumps: antibiotics are exported via the membrane proteins inside the cell and maintain their low-intracellular concentrations by pumping them out before they reach their target. Several unrelated antibiotics such as macrolides, tetracyclines, and fluoroquinolones (FQ) are been pumped by cytoplasmic membrane-associated multidrug transporters contributing in the development of MDR strains. ❸ Inhibitors of DNA replication: inhibition of the bacterial division by preventing enzyme DNA gyrase activity (nicks double-stranded DNA and negative supercoils and reseals) is facilitated by quinolones like FQ. ❹ Altered cell wall precursors: Gram-positive bacteria can be inhibited by glycopeptides like vancomycin or teicoplanin as they are involved suppression cell wall synthesis. ❺ DNA mutated-DNA gyrase and topoisomerase IV: quinolone mechanism of resistance involves modification of two enzymes: DNA gyrase and topoisomerase IV. Replication failure is due to mutations in genes gyrA and parC making FQ unable to bind. ❻ Folic acid metabolism inhibitors: combination of few drugs like sulphonamides and trimethoprim acts on biosynthetic pathway, which shows synergy and a reduced mutation rate for resistance. ❼ Alteration in the 30S subunit or 50S subunit: resistance to drugs that affect protein synthesis, that is, macrolides, tetracycline, and chloramphenicol, bind to 30S ribosomal subunit, whereas chloramphenicol, macrolides, lincosamides, and streptogramin B bind to 50S ribosomal subunit to suppress protein synthesis. ❽ Inhibitors of protein synthesis: tetracyclines offer resistance by inhibiting integrity of ribosomal structure and RNA polymerase mutations conferring resistance to rifampicin. ❾ Antibiotic inactivation by enzymes: β-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferases are the three main enzymes that inactivate antibiotics. ❿ Porins: present in the outer membrane. The changed selectivity or decreased in porins stops the antimicrobials to act on the pathogens conferring resistance.
