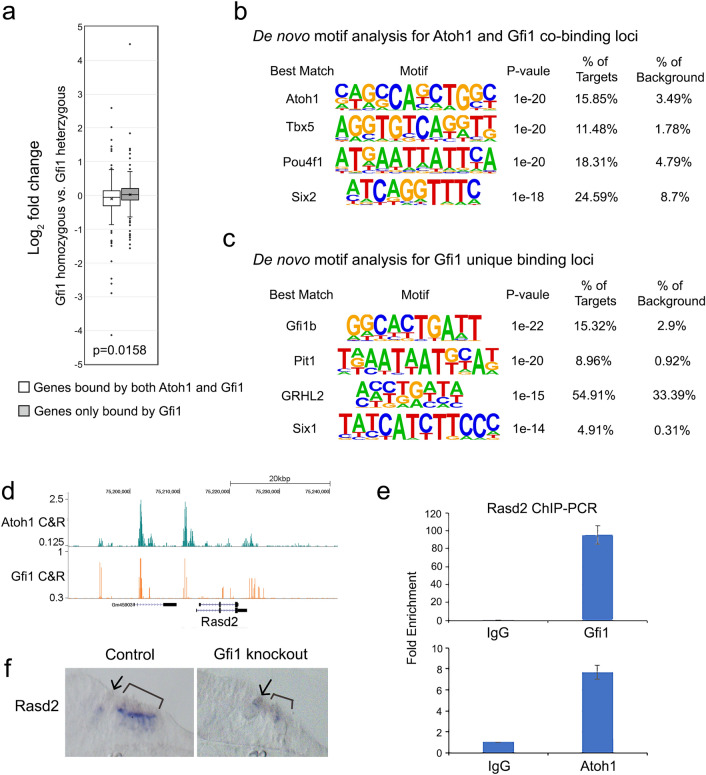
Figure 5.
GFI1 positively regulate many hair cell genes without evidence for direct binding to DNA. (a) Box plot showing that genes bound by both ATOH1 and GFI1 are more likely to be down-regulated in Gfi1 mutant hair cells (i.e. positively regulated by GFI1), while genes bound by only GFI1 are more likely to be up-regulated in Gfi1 mutant hair cells (i.e. negatively regulated by GFI1). Statistical significance was assessed by a Student t-test, comparing the log2 fold change (FC) values. (b) De novo motif analysis of the 386 loci bound by both ATOH1 and GFI1. Although ATOH1 DNA recognition motifs are highly enriched in these peaks, Gfi1 DNA recognition motifs are not. (c) De novo motif analysis of the 350 loci bound by only by GFI1. Here, the most highly enriched motif is a GFI1b recognition motif. (d) Genomic browser track of CUT&RUN sequencing showing both ATOH1 and GFI1 bind to loci near the hair cell gene Rasd2. (e) ChIP-QPCR analysis of ATOH1 and GFI1 binding to the same distal regulatory elements in the Rasd2 gene. Cochlear epithelium from neonatal Atoh1-GFP mice were processed for ChIP using antibodies for GFP (to pull down ATOH1-bound DNA) and GFI1, with IgG as a negative control. (f) In situ hybridization of Rasd2 mRNA in P0 Gfi1 wild type and mutant cochlea. Arrow marks inner hair cells, brackets mark the outer hair cell region.