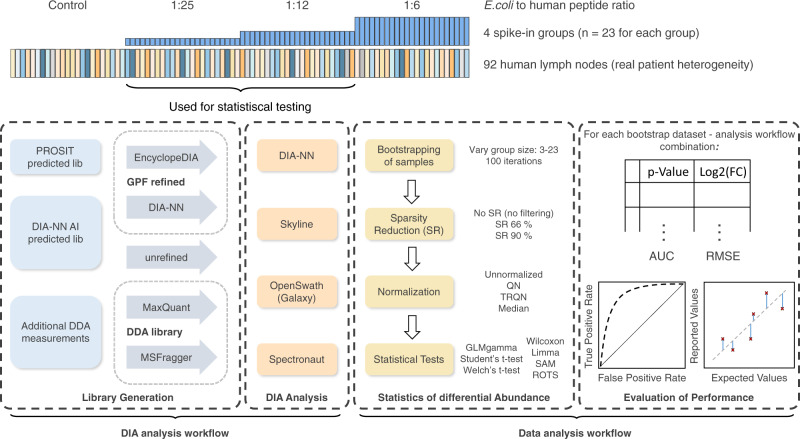
Fig. 1. Benchmarking workflow.
A data-independent acquisition (DIA) benchmark dataset was created by adding E. coli peptides in known ratios to peptide preparations of lymph nodes of 92 individuals. We analyzed the raw data with different spectral libraries and DIA software suites. From samples to which E. coli peptides were added in the two E. coli: human peptide ratios 1:25 and 1:12, bootstrap datasets with group sizes of 3 to 23 were generated. For each of those 21 different group sizes, 100 bootstrap datasets were generated. On each bootstrap dataset different data analysis workflows, composed of different sparsity reductions, normalization options, and different statistical tests for detecting differentially abundant proteins, were applied. The results were returned in a table containing p-values and log2 fold-changes (log2FCs) for each protein. As the ground truth about the changed proteins (E. coli) is known, the prediction performance of each workflow can be assessed. This can be done based on the p-values from the statistical tests by calculating the receiver operating characteristic (ROC) curve, based on which the area under curve (AUC) is calculated. To quantify the accuracy of quantification the root-mean-square error (RMSE) is calculated based on the detected log2FC.