Abstract
Background. Uterine leiomyosarcoma is a rare aggressive smooth muscle cancer with poor survival rates. RNA Polymerase I (Pol I) activity is elevated in many cancers supporting tumour growth and prior studies in uterine leiomyosarcoma revealed enlarged nucleoli and upregulated Pol I activity-related genes. This study aimed to investigate the anti-tumour potential of CX-5461, a Pol I transcription inhibitor currently being evaluated in clinical trials for several cancers, against the human uterine leiomyosarcoma cell line, SK-UT-1. Methods. SK-UT-1 was characterised using genome profiling and western blotting. The anti-tumour effects of CX-5461 were investigated using cell proliferation assays, expression analysis using qRT-PCR, and BrdU/PI based cell cycle analysis. Results. Genetic analysis of SK-UT-1 revealed mutations in TP53, RB1, PTEN, APC and TSC1 & 2, all potentially associated with increased Pol I activity. Protein expression analysis showed dysregulated p53, RB1 and c-Myc. CX-5461 treatment resulted in an anti-proliferation response, G2 phase cell-cycle arrest and on-target activity demonstrated by reduced ribosomal DNA transcription. Conclusions. SK-UT-1 was confirmed as a representative model of uterine leiomyosarcoma and CX-5461 has significant potential as a novel adjuvant for this rare cancer.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10637-022-01222-w.
Keywords: Uterine leiomyosarcoma, SK-UT-1, RNA Polymerase I transcription inhibitor, CX-5461
Introduction
Uterine leiomyosarcoma is a subtype of soft tissue sarcoma and arises in the smooth muscle of the uterus. Although rare, accounting for < 5% of female genital tract cancers, and the fact that 60% of cases are diagnosed at an early stage, it is an extremely aggressive and progressive cancer with a 5-year survival rate for localised disease of 52% and metastatic disease rate of 12% [1, 2]. Surgical resection and chemotherapy are used as the mainstay for uterine leiomyosarcoma management, however, the curative effects are limited [3–5]. In addition, loss of heterozygosity at loci containing the tumour suppressor genes, tumour protein 53 (TP53), retinoblastoma 1 (RB1), phosphatase and tensin homolog (PTEN) and cyclin-dependent kinase inhibitor protein (CDKN2A), in uterine leiomyosarcoma confers chemotherapy resistance [6, 7]. There is an urgent need, therefore, to identify new targets and effective therapeutic strategies to improve the outcomes of this aggressive disease.
Rapid growth of cancers, one of the classic hallmarks, is associated with elevated rates of protein synthesis and ribosome biogenesis (RiBi) [8]. An early rate limiting step in RiBi, is the transcription of the 47S precursor ribosomal RNA (rRNA) by Pol I, which is rapidly processed into mature rRNA forming the nucleic backbone of ribosomes. RiBi occurs in the nucleolus at elevated levels in cancer cells to sustain the rapid growth rate [9, 10]. Enhanced Pol I transcription activity can be mediated by altered signalling pathways and genetic mutations in key oncogenes or tumour suppressors (PI3K/Akt, RAS/MAPK, mTOR, MYC, P53, RB1 and ATRX). It is for these reasons Pol I was considered a key target for cancer treatment and specific inhibitors were developed [11].
Genetic studies of uterine leiomyosarcoma tumours from patients report that genes related to Pol I transcription activity, including TP53, RB1, ATRX and MYC, are frequently mutated. TP53 and RB1 were confirmed as the most frequently altered genes with 61% and 48%, respectively, showing somatic mutations or homozygous deletions. ATRX was mutated in 34% and MYC was amplified in 38% of cases [7, 12]. Enlarged nucleoli, a marker of aggressive disease, and increased rDNA transcription have also been reported in uterine leiomyosarcoma [13]. Taken together, these studies suggest that inhibition of Pol I transcription may be an effective new therapeutic strategy for the treatment of uterine leiomyosarcoma. To investigate this hypothesis, (i) we analysed the genetic profile of the human uterine leiomyosarcoma cell line, SK-UT-1, to identify key driver mutations linked to Pol I activity and to establish the cell line as a representative in vitro model of this rare cancer type, hence developing a robust and suitable model system for testing novel therapies, and (ii), we evaluated the effects of the Pol I transcription inhibitor, CX-5461 [14–16], against this cell line.
Methods
Genomic profiling of SK-UT-1
The Ampliseq™ Cancer Childhood Panel DNA assay (Illumina) detects single nucleotide variants (SNV) from hotspots of 86 genes, full exons of 44 genes, and copy number variants (CNV) from 28 clinically relevant cancer genes. DNA was extracted from the cell lines using ReliaPrep tissue DNA extraction kit (Promega, Madison, WI, USA). The Childhood Cancer Research Assay primers and AmpliSeq Library Kit Plus (Illumina) were used for library preparation. The prepared libraries were sequenced using a MiniSeq sequencer (Illumina). Base calling and mapping to a reference genome (hg19) was performed using the BaseSpace Informatics suite (Illumina). SNV variant calling was performed in DNA amplicon application (Illumina, version 2.1.1). CNV calling was performed in OncoCNV caller application (Illumina, version 1.2.0). All VCF files were loaded into variant interpreter (version 2.7.0.412) for interpretation. SNV somatic candidates were selected based on minimum coverage reads of more than 100, minimum allele frequency of 10%, cosmic reported variant and less than 1% prevalence in the 1000 Genome Population Database. CNV candidates with more than 10 gene copies were identified.
Cell culture and IncuCyte-based cell proliferation assay
Human uterine leiomyosarcoma cell line, SK-UT-1, was purchased from American Tissue Culture Collection (ATCC®, Manassas, VA, USA) and cultured with minimum essential medium (MEM) containing 10% Foetal Bovine Serum (FBS) (Sigma-Aldrich, St Louis, MO), 2 mM L-glutamine (Gibco, Waltham, MA), 1% Puromycin/Streptomycin/Neomycin (Gibco, Waltham, MA). 1 × 103 of SK-UT-1 cells were seeded into a 96-well plate and incubated for 12 h (hr). The medium was then exchanged with fresh complete medium containing vehicle or various concentrations of CX-5461 and incubated for 72 h in a ZOOM IncuCyte® Live Cell Imaging System. The confluence in each well was recorded every 12 h. The 50% growth inhibitory concentration (GIC50) of CX-5461 was determined by Graph-Pad Prism (Version 9.1.0). A human foreskin fibroblast cell line was purchased from Lonza (Lonza, Basel, Switzerland). The human foreskin fibroblast cells were cultured with Dulbecco’s modified eagle medium (DMEM) containing 10% of FBS, 8 mM L-glutamine, 1% Puromycin/Streptomycin/Neomycin.
Western blotting
SK-UT-1 cells (1.0 × 106) and human fibroblast cells (1.0 × 106) were harvested, washed, then lysed in cell lysis buffer containing 0.5 mM EDTA, 2% SDS, 20 mM HEPES and protease inhibitor (Roche, Basel, Switzerland). The protein concentration was measured using a DC™ protein assay kit (Bio-Rad, Hercules, CA) and 30 µg of protein was resolved by SDS-PAGE. Once separated, the proteins were transferred to a Polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Hercules, CA) using the Bio-Rad semi-dry transfer protocol (1 mA current, 30 V constant voltage for 30 min). PVDF membranes were rinsed in 1X Tris-Buffer Saline Tween-20 (TBST) (1X TBS (pH7.5) 1 mM Tris, 10 mM NaCl, 0.1% Tween 20), then blocked with 5% non-fat dry milk in 1X TBST for 1 h at room temperature. The membrane was probed with primary antibodies (Supplementary Table 2), diluted in 5% BSA in 1X TBST, overnight at 4 °C then washed 1X TBST and incubated with horseradish peroxidase-conjugated (HRP) secondary antibodies (Supplementary Table 2) for 1 h at room temperature. The membranes were washed with 1X TBST, developed for enhanced chemiluminescence using Clarity Western ECL Substrate reagent (Bio-Rad, Hercules, CA), then the blots were imaged with a ChemiDoc MP Imager (Bio-Rad, Hercules, CA). β-actin was used as a loading control. Band intensity was analysed using Bio-Rad Image Lab Software (Version 6.0.1).
Quantitative Real-Time PCR for measurement of rDNA transcription
rDNA transcription was determined by qPCR as previously described Bywater et al. [15]. Briefly, SK-UT-1 cells (1.0 × 105) were seeded into 100 mm cell culture dishes and incubated for 12 h, following which the medium was exchanged with fresh complete medium containing vehicle or various concentrations of CX-5461, then incubated for 1 h. RNA was then extracted using a RNeasy® Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA (500 ng) was DNase treated (according to kit instructions), then incubated in T100™ Thermocycler (Bio-Rad, CA, USA) using the settings detailed in Supplementary Table 3. cDNA synthesis was performed using SuperScript™ IV Reverse Transcriptase (Invitrogen, Carlsbad, CA), Hexamer Random Primers (Promega, Madison, WI), and dNTPs (Invitrogen, Carlsbad, CA) with incubation in a T100™ Thermocycler (detailed in Supplementary Table 3). RT-PCR was performed using SYBR™ Green master mix (Applied Biosystems, Foster City, CA) and primers (Supplementary Table 4) in a StepOne™ Plus Real Time (RT) PCR System (Applied Biosystems) with the settings detailed in Supplementary Table 3.
BrdU and PI staining for cell cycle analysis
Bromodeoxyuridine (BrdU) and propidium iodide (PI) staining assay was performed following the manufacturer’s instructions. Briefly, 3 × 104 cells were seeded in 100 mm cell culture dish and incubated for 12 h. The media was then changed to complete medium containing vehicle or CX-5461 at the GIC50 concentrations determined in the cell proliferation assay. After 72 h, 10 µM BrdU (Sigma-Aldrich, St Louis, MO) was added for 30 min and the cells harvested and stained with the anti BrdU specific antibody (Clone: B44, BD Bioscience, Franklin Lakes, NJ) and PI (Sigma Aldrich, St Louis, MO) according to the manufacturer’s instructions. Following the staining, samples were analysed using flow cytometry on a Becton Dickinson LSR II FACS machine and the data analysed using FlowJo software (Version 10.0).
Statistical analysis
Statistical analyses were conducted using GraphPad Prism software (Version 9.1.0). Comparison of western blotting band intensities between SK-UT-1 and human fibroblast (HF) cell lines was conducted using a Mann–Whitney U test. Statistical significance between two groups was assessed by two-way ANOVA. Dose–response curve fit and 50% inhibitory values were determined using non-linear regression analysis.
Results
Pol I transcription-related genes are altered in the SK-UT-1 cell line
Genomic profiling of the SK-UT-1 cell line revealed missense, non-sense, frameshift and splice-site mutations in 21 genes (Supplementary Table 1). Consistent with prior uterine leiomyosarcoma genomic studies [7, 12], TP53, RB1 and PTEN mutations were identified in the SK-UT-1 cells. Mutations were also found in Phosphatidylinositol 3-kinase (PIK3CA) and Adenomatous polyposis coli protein (APC) genes, that are reported to impact Pol I transcription activity [9, 10]. Deleterious mutations in the tumour suppressor genes, Tuberous Sclerosis Complex 1 (TSC1) and TSC2, were also identified which have not been previously reported to our knowledge. Western blotting analysis of protein lysate generated from SK-UT-1 cells demonstrated elevated levels of p53 and c-Myc protein, but undetectable levels of RB1 protein in comparison to the human fibroblast (HF) control cell line (Fig. 1a - c). These mutations were consistent with the expected sequelae of TP53 gain of function hotspot mutations, R248Q and R175H, leading to stabilisation of p53, and RB1 frameshift variant detected [17–19].
Fig. 1.
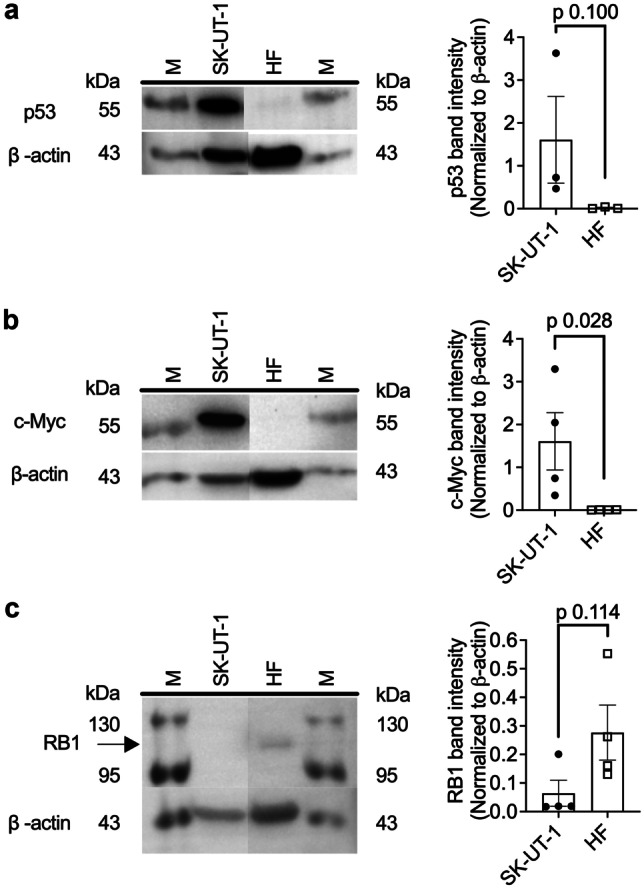
Basal expression of key proteins on uterine leiomyosarcoma SK-UT-1 cells. Basal protein abundance of (a) p53, (b) c-Myc and (c) RB1 were evaluated by western blot analysis. Human fibroblasts (HF) were used as a control cell line (note: intervening lanes with non-relevant cell lines have been removed from the image). M is abbreviated for marker lane. Band intensities were normalized to β-actin. Statistical analysis was conducted using a Mann Whitney U test (Error bars represent mean ± SEM of n = 3–4 biological replicates)
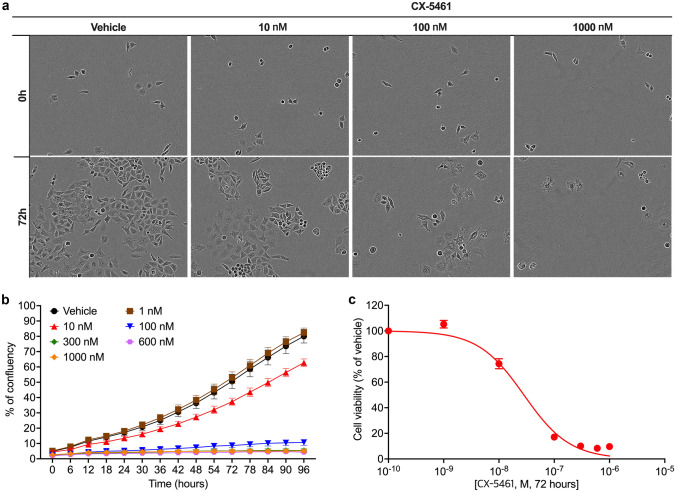
CX-5461 inhibits uterine leiomyosarcoma cell proliferation
The antitumour potential of Pol I transcription inhibitor CX-5461 on SK-UT-1 was assessed by a cell proliferation assay (Fig. 2). Treatment studies with CX-5461 demonstrated a dose-dependent decrease in cell density (% of confluency) following 72 h of drug exposure (Fig. 2a, b). The 50% growth inhibitory concentration (GIC50) was calculated at 28.1 ± 3.1 nM (Fig. 2c).
Fig. 2.
Anti-proliferation effects of CX-5461 on SK-UT-1 cells. (a) SK-UT-1 cells were treated with CX-5461, at the concentrations indicated, or vehicle (50 mM NaH2PO4 (pH4.5)) and assessed for proliferation via phase contrast microscopy and IncuCyte® ZOOM Live Cell Imaging System (Error bars represent mean ± SEM of n = 3 biological triplicates). A dose–response curve (c), generated from data obtained in (b), with the 50% growth inhibitory concentration (GIC50) of CX-5461 calculated using non-linear regression analysis
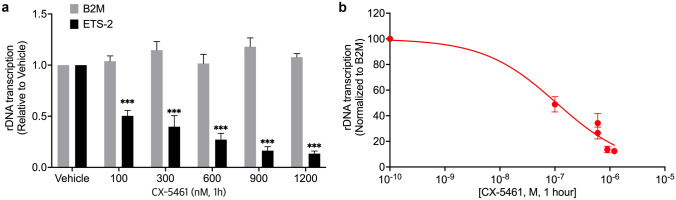
rDNA transcription is targeted by CX-5461
To investigate the effect of CX-5461 on its primary target, rDNA transcription, the abundance of 5’-External Transcribed Spacer (ETS), a rapidly processed region of the 47S pre-ribosomal RNA, was measured by q-RT-PCR after CX-5461 treatment (1 h). CX-5461 effectively inhibited rDNA transcription rate in a dose-dependent manner (Fig. 3a). The 50% transcription inhibitory concentration (tIC50) for CX-5461 was calculated at 112 ± 32 nM (Fig. 3b).
Fig. 3.
Inhibitory effects of CX-5461 on rDNA transcription in SK-UT-1 cells (a) The effects of treatment with CX-5461 at varying concentrations, and vehicle control, for 1 h on rDNA transcription rate in SK-UT-1 cells, determined via qPCR with the result of the external transcribed spacer (ETS) normalised to the house-keeping gene, β2 microglobulin (β2M). Statistically significance differences between ETS-2 and β2M were determined using two-way ANOVA (Error bars represent mean ± SEM of n = 3) *p <0.05, **p <0.01, ***p <0.001. (b) The 50% rDNA transcription inhibitory concentration (tIC50) of CX-5461 was calculated from (a) using non-linear regression analysis (Error bars represent mean ± SEM of n = 3 biological triplicates)
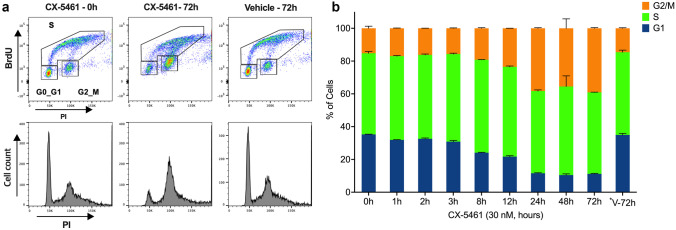
CX-5461 induces G2 phase cell cycle arrest
SK-UT-1 was identified as p53 mutant in the genetic analysis and protein expression assays (Supplementary Table 1 and Fig. 1a). In the absence of wild-type p53, CX-5461 has previously been demonstrated to induce cell cycle defects that result in the accumulation of cells in the G2 phase of the cell cycle [20]. To investigate the effect of CX-5461 on SK-UT-1 cell cycle progression, BrdU and PI staining assays were conducted (Fig. 4a). Consistent with previous studies, cell cycle analysis revealed an increase in the percentage of cells in G2/M phase of the cell cycle and a reduction in the G1 subpopulation after 8 h of CX-5461 treatment (Fig. 4b). No change was detected in the percentage of cells in S phase at the treatment timepoints analysed.
Fig. 4.
Effects of CX-5461 on cell cycle progression in SK-UT-1 cells (a) Flow cytometry gating strategy for cell cycle analysis using BrdU and PI staining at 0 and 72 h following treatment with 30 nM CX-5461, or vehicle, is shown. (b) SK-UT-1 cells were treated with the GIC50 CX-5461 dose (30 nM) for the times indicated, and stained with a anti BrdU specific antibody and PI for cell cycle analysis as in (a) (bars represent mean ± SD of n = 2 biological duplicates). *V-72 h: Vehicle for 72 h
Discussion
This paper details the first studies evaluating the therapeutic potential of the Pol I transcription inhibitor, CX-5461, against uterine leiomyosarcoma. CX-5461, a first-in-class Pol I transcription inhibitor, selectively inhibits Pol I transcription by preventing pre-initiation complex formation at the rDNA promoter, and has demonstrated broad-spectrum anticancer therapeutic efficacy in multiple in vitro and in vivo studies of human cancers [21] (lymphoma [15], AML [22], neuroblastoma [23], breast cancer [24], osteosarcoma [25] and prostate cancer [26]). CX-5461 was well-tolerated in a recently completed phase I clinical trial in patients with haematological malignancy [16], and a phase II clinical trial for breast cancer (NCT02719977) is currently underway. Recently, Sanij et al.reported the therapeutic potential of CX-5461 in pre-clinical studies of ovarian cancer [27], however, the effects of CX-5461 on uterine leiomyosarcoma have not yet been evaluated.
Genes that influence Pol I transcription activity, such as TP53, RB1, PTEN, ATRX and MYC, have been found to be mutated in uterine leiomyosarcoma tumours, with TP53 and RB1 being the most-commonly altered genes [7, 12]. MYC regulates rDNA transcription through two mechanisms; firstly it directly transcriptionally upregulates the majority of the Pol I components [28]; secondly, it binds to rDNA promoter regions and promotes Pol I transcription through chromatin remodelling and interaction with Pol I-related cofactor [29]. In contrast, p53 and RB suppress Pol I transcription by disrupting Pol I binding to the rDNA gene promoter [30, 31].
Consistent with prior analysis [32, 33], our genomic profiling of the SK-UT-1 uterine leiomyosarcoma cell line confirmed mutations in TP53, RB1, APC and PTEN, but also the tumour suppressors TSC1 and TSC2. TSC1 and TSC2, together with the auxiliary subunit, Tre-Bud-Cdc16-1 domain member 7 (TBC1D), form the TSC complex [34] which regulates cell growth by controlling mTORC1 activation [35]. Loss of either TSC1 or TSC2 also increases c-Myc expression [35, 36], most likely as a direct result of mTORC1 signalling activation [37]. This would, in turn, enhance Pol I transcription activity [27, 38]. Evidence supporting a direct role for the TSC complex in this cancer type is that the allelic loss of TSC2 in Eker mutant rats results in the spontaneous development of uterine leiomyosarcoma [39]. Western blotting studies in SK-UT-1 found elevated levels of p53 and c-Myc, and undetectable levels of RB1. In combination, these findings demonstrate that the SK-UT-1 cell line is broadly representative of uterine leiomyosarcoma tumours, and supports the likelihood that Pol I inhibition would be effective against this cancer type.
The major consequence of inhibiting rDNA transcription is activation of the nucleolar stress pathway through p53-dependent and p53-independent pathways [15, 20, 40]. CX-5461 has previously been shown to (i) activate a p53-dependent G1 phase arrest, in which cells may undergo apoptosis, cell cycle arrest, differentiation and/or senescence, and (ii) a p53-independent DNA damage response (DDR) [24, 27, 41]. Additionally, activation of the Ataxia telangiectasia mutated (ATM) and Ataxia telangiectasia and Rad3 related (ATR) kinase pathways by CX-5461 mediates a G2 phase cell cycle arrest [20]. In our studies, CX-5461 rapidly reduced the rate of rDNA transcription in p53 mutant SK-UT-1 cells following 1 h of treatment, with subsequent anti-proliferative effects observed from 12 h of treatment, presumably mediated via the observed G2 phase cell cycle arrest noted by 8 h of treatment. Relatively low nanomolar CX-5461 concentrations were required to achieve these effects with the GI50 and tIC50 values being similar to those reported for other sensitive cancer cell lines [15, 27]. Dose escalation and pharmacokinetic/dynamic studies of CX-5461 in humans with haematological malignancies revealed that, even at the lowest dose of 25 mg/m2, Pol I inhibition occurred in tumour cells [16]. The striking proliferation defect at low nanomolar concentrations in our vitro studies (tIC50 = 112 ± 32 nM) are within the same range as the concentrations obtained in vivo [16], thus suggesting that the drug concentrations used in this study are translatable to therapeutically effective in vivo concentrations.
In conclusion, these studies (i) confirm the SK-UT-1 cell line as a representative model of uterine leiomyosarcoma, and (ii) demonstrate that targeting dysregulated Pol I transcription in this cancer type with CX-5461 has significant potential as a novel adjunct therapy. To strengthen these findings, future studies could include additional uterine leiomyosarcoma cell lines and in vivo mouse models of uterine leiomyosarcoma. However, given the poor outcomes of this disease with current treatment, the targeted biological mechanism being common to many cancers, and the in-human experience already gained with CX-5461 against several other cancer types, small pilot trials of CX-5461 in combination with salvage chemotherapy could be justified in patients who have failed standard treatment regimes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Thanks to the staff of Imaging and Cytometry and the ACRF Biomolecular Resource Facilities of The John Curtin School of Medical Research, The Australian National University, for their assistance with equipment and settings.
Author’s contributions
C-WK performed experiments, analysed data and prepared manuscript; KMH and ACB planned and supervised experiments, analysed data and edited manuscript; ALHP, KCH and GJY funded, planned, undertook and analysed genetic profiling of SK-UT-1 cell line and contributed to manuscript; NH assisted with planning and execution of experiments; DD intellectual input; RDH intellectual and financial input; LAC part-funded and managed the project, planned and supervised experiments, analysed data and edited manuscript.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported through kind donations made by Dr. David Boyle and family in memory of his wife Donna, and by Dr Grace Moshi of the Sarah Grace Sarcoma Foundation. Additionally, a National Health and Medical Research (NHMRC) of Australia Program Grant (APP No.1053792) supported this work and a NHMRC Principal Research Fellowship (No. 116999).
Data availability
Data will be made available upon request through the Corresponding Author.
Declarations
Ethics approval
Not applicable.
Informed consent
This study does not contain any individual person’s data.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Research involving human participants and/or animals
This study does not involve human participants, human data or animal data.
Competing interests
The authors declare no potential conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Seagle BL, et al. Prognosis and treatment of uterine leiomyosarcoma: A National Cancer Database study. Gynecol Oncol. 2017;145(1):61–70. doi: 10.1016/j.ygyno.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Hoang HL et al (2014) Prognostic factors and survival in patients treated surgically for recurrent metastatic uterine leiomyosarcoma. Int J Surg Oncol 2014:919323 [DOI] [PMC free article] [PubMed]
- 3.Gadducci A, et al. Uterine leiomyosarcoma: analysis of treatment failures and survival. Gynecol Oncol. 1996;62(1):25–32. doi: 10.1006/gyno.1996.0185. [DOI] [PubMed] [Google Scholar]
- 4.Pautier P et al (2013) A randomized clinical trial of adjuvant chemotherapy with doxorubicin, ifosfamide, and cisplatin followed by radiotherapy versus radiotherapy alone in patients with localized uterine sarcomas (SARCGYN study) A study of the French Sarcoma Group. Ann Oncol 24(4):1099–104 [DOI] [PubMed]
- 5.Hensley ML et al (2013) Adjuvant therapy for high-grade, uterus-limited leiomyosarcoma: results of a phase 2 trial (SARC 005). Cancer 119(8):1555–61 [DOI] [PubMed]
- 6.Zhai YL, et al. Frequent occurrence of loss of heterozygosity among tumor suppressor genes in uterine leiomyosarcoma. Gynecol Oncol. 1999;75(3):453–459. doi: 10.1006/gyno.1999.5629. [DOI] [PubMed] [Google Scholar]
- 7.Astolfi A et al (2020) Genomic Database Analysis of Uterine Leiomyosarcoma Mutational Profile Cancers. (Basel) 12(8) [DOI] [PMC free article] [PubMed]
- 8.Hanahan D and RA Weinberg (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–74 [DOI] [PubMed]
- 9.Drygin D. WG Rice and I Grummt. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier J, Thomas G, Volarevic S (2018) Ribosome biogenesis in cancer: new players and therapeutic avenues .Nat Rev Cancer 18(1):51–63 [DOI] [PubMed]
- 11.Ferreira R et al (2020) Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 9(2) [DOI] [PMC free article] [PubMed]
- 12.Cuppens T, et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways. Int J Cancer. 2018;142(6):1230–1243. doi: 10.1002/ijc.31129. [DOI] [PubMed] [Google Scholar]
- 13.Avdalyan A et al (2012) Prognostic Value of Microvessel Density in Tumor and Peritumoral Area as Evaluated by CD31 Protein Expression and Argyrophilic Nucleolar Organizer Region Count in Endothelial Cells in Uterine Leiomyosarcoma. Sarcoma 2012:594512 [DOI] [PMC free article] [PubMed]
- 14.Drygin D et al (2011) Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 71(4):1418–30 [DOI] [PubMed]
- 15.Bywater MJ et al (2012) Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53 Cancer. Cell 22(1):51–65 [DOI] [PMC free article] [PubMed]
- 16.Khot A et al (2019) First-in-Human RNA Polymerase I Transcription Inhibitor CX-5461 in Patients with Advanced Hematologic Cancers: Results of a Phase I Dose-Escalation. Study Cancer Discov 9(8):1036–1049 [DOI] [PubMed]
- 17.Hernández-Reséndiz I et al (2019) Mutant p53. J Cell Physiol 234(5):5524–5536 [DOI] [PubMed]
- 18.Senapati P et al (2018) Oncogene c-fos and mutant R175H p53 regulate expression of Nucleophosmin implicating cancer manifestation. FEBS J 285(18):3503–3524 [DOI] [PubMed]
- 19.Yan W, Chen X (2009) Identification of GRO1 as a critical determinant for mutant p53 gain of function. J Biol Chem 284(18):12178–87 [DOI] [PMC free article] [PubMed]
- 20.Quin J et al (2016) Inhibition of RNA polymerase I transcription initiation by CX-5461 activates non-canonical ATM/ATR signaling. Oncotarget 7(31):49800–49818 [DOI] [PMC free article] [PubMed]
- 21.Haddach M et al (2012) Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics ACS. Med Chem Lett 3(7):602–6 [DOI] [PMC free article] [PubMed]
- 22.Hein N, et al. Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population. Blood. 2017;129(21):2882–2895. doi: 10.1182/blood-2016-05-718171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hald Ø, et al. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene. 2019;38(15):2800–2813. doi: 10.1038/s41388-018-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H et al (2017) CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun 8:14432 [DOI] [PMC free article] [PubMed]
- 25.Li L et al (2016) CX-5461 induces autophagy and inhibits tumor growth via mammalian target of rapamycin-related signaling pathways in osteosarcoma. Onco Targets Ther 9:5985–5997 [DOI] [PMC free article] [PubMed]
- 26.Rebello RJ, et al. The Dual Inhibition of RNA Pol I Transcription and PIM Kinase as a New Therapeutic Approach to Treat Advanced Prostate Cancer. Clin Cancer Res. 2016;22(22):5539–5552. doi: 10.1158/1078-0432.CCR-16-0124. [DOI] [PubMed] [Google Scholar]
- 27.Sanij E et al (2020) CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat Commun 11(1):2641 [DOI] [PMC free article] [PubMed]
- 28.Poortinga G et al (2011) c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res 39(8):3267–81 [DOI] [PMC free article] [PubMed]
- 29.Poortinga G, Quinn LM, Hannan RD (2015) Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene 34(4):403–12 [DOI] [PubMed]
- 30.Zhai W, Comai L (2000) Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol 20(16):5930–8 [DOI] [PMC free article] [PubMed]
- 31.Hannan KM et al (2000) Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 19(43):4988–99 [DOI] [PubMed]
- 32.Tate JG et al (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47(D1):D941-D947 [DOI] [PMC free article] [PubMed]
- 33.Ghandi M, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H et al (2021) Structural insights into TSC complex assembly and GAP activity on Rheb. Nat Commun 12(1):339 [DOI] [PMC free article] [PubMed]
- 35.Schmidt EV, et al. Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway. Cell Cycle. 2009;8(9):1344–1351. doi: 10.4161/cc.8.9.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun S et al (2015) Constitutive Activation of mTORC1 in Endothelial Cells Leads to the Development and Progression of Lymphangiosarcoma through VEGF Autocrine Signaling Cancer. Cell 28(6):758–772 [DOI] [PMC free article] [PubMed]
- 37.Al-Saleem T et al (1998) Malignant tumors of the kidney, brain, and soft tissues in children and young adults with the tuberous sclerosis complex. Cancer 83(10):2208–16 [PubMed]
- 38.Hannan KM et al (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23(23):8862–77 [DOI] [PMC free article] [PubMed]
- 39.Kubo Y et al (1995) Allelic loss at the tuberous sclerosis (Tsc2) gene locus in spontaneous uterine leiomyosarcomas and pituitary adenomas in the Eker rat model. Jpn J Cancer Res 86(9):828–32 [DOI] [PMC free article] [PubMed]
- 40.Donati G, Montanaro L, Derenzini M (2012) Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res 72(7):1602–7 [DOI] [PubMed]
- 41.El Hassouni B et al (2019) CX-5461 Inhibits Pancreatic Ductal Adenocarcinoma Cell Growth, Migration and Induces DNA Damage. Molecules 24(24) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request through the Corresponding Author.