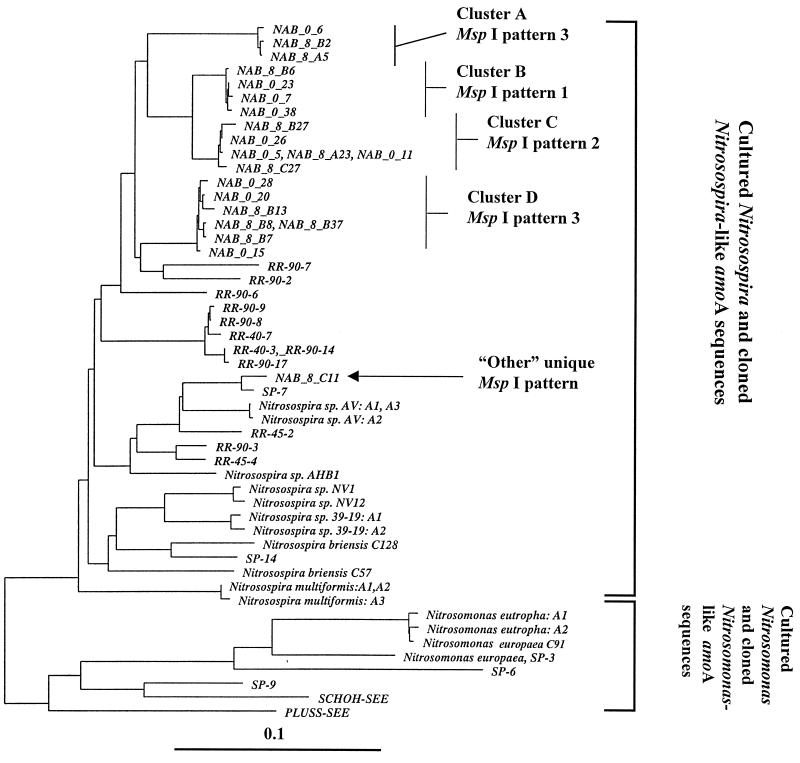
FIG. 3.
Neighbor-joining tree with Fitch-Margoliash correction of amoA sequences recovered from soil microcosms. The tree is derived from a compilation of available amoA sequences spanning the 449 nucleotide bases available for all sequences used. Sequences prefixed with NAB were generated during this study (accession no. AF056049 to AF056069). Clones were selected from libraries on the basis of MspI restriction patterns to provide a survey of sequence diversity in the target amoA sequences. Sequences which are identical over the fragment analyzed are presented on the same branch and separated by commas. Reference sequences: sequences SCHOH-SEE and PLUSS-SEE and sequences prefixed with RR and SP (environmental clones) (33); Nitrosospira briensis C57, Nitrosovibrio tenuis NV1, and Nitrosomonas europaea C-91 (pure cultures) (33); Nitrosospira sp. strain AHB1 (32); Nitrosomonas eutropha copies A1 and A2: (accession no. U51630 and U72670) (25); Nitrosospira multiformis copies A1, A2 and A3 (accession no. U91603, U15733, and U89833 respectively) (25); Nitrosospira briensis C128 (accession no. U76553) (25); Nitrosovibrio tenuis NV12 (accession no. U76552) (25); Nitrosospira sp. strain 39-19 copies A2 and A3 (accession no. AF016002 and AF006692, respectively) (25); Nitrosomonas europaea (23).