Figure 9. Bacterial detection with PAD.
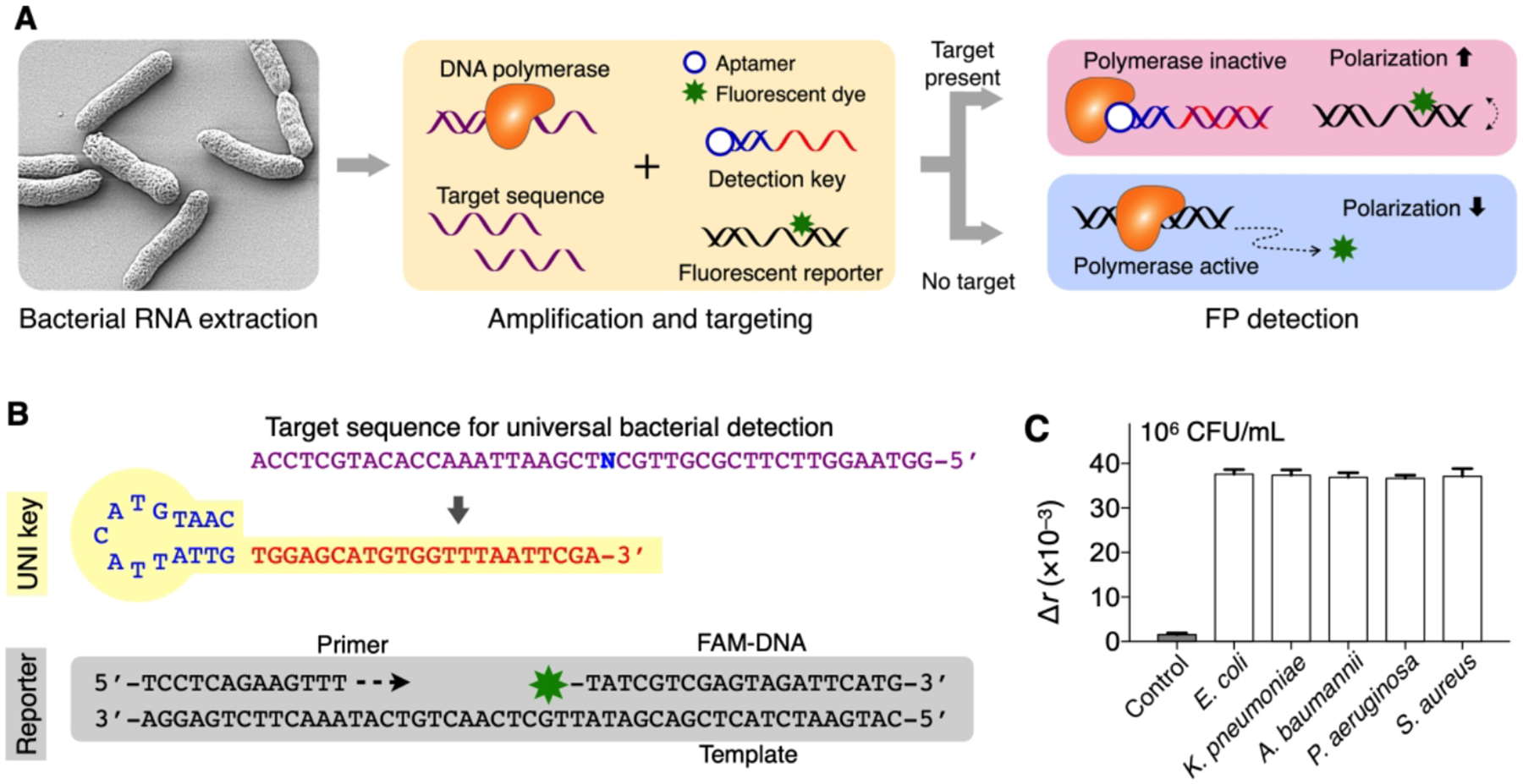
(A) Assay principle. Following the extraction and amplification of target NAs, the sample is mixed with a dual probe set. In the presence of target NA, the detection key is stabilized through hybridization and locks into DNA polymerase, which deactivates polymerase activity. The fluorescent reporter DNA then retains its structure and assumes high fluorescence anisotropy (r). In the absence of target NA, unlocked DNA polymerase cleaves the reporter’s fluorophore during the extension reaction, which leads to low r values. (B) Universal bacteria detection with UNI key. A conserved sequence was targeted (N = A for Escherichia, Klebsiella, Acinetobacter; N = T for Pseudomonas, Staphylococcus). (C) Five different HAI pathogens (106 CFU/mL) were detected. The signals were statistically identical (P = 0.886, one-way ANOVA) among concentration-matched samples. The bar graphs display mean ± s.d. (triplicates). Adapted with permission from ref 3. Copyright 2016 © The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science.
