Figure 2. Deconvolved zero-charge mass spectra of the purified M. maripaludis RNase P subunits.
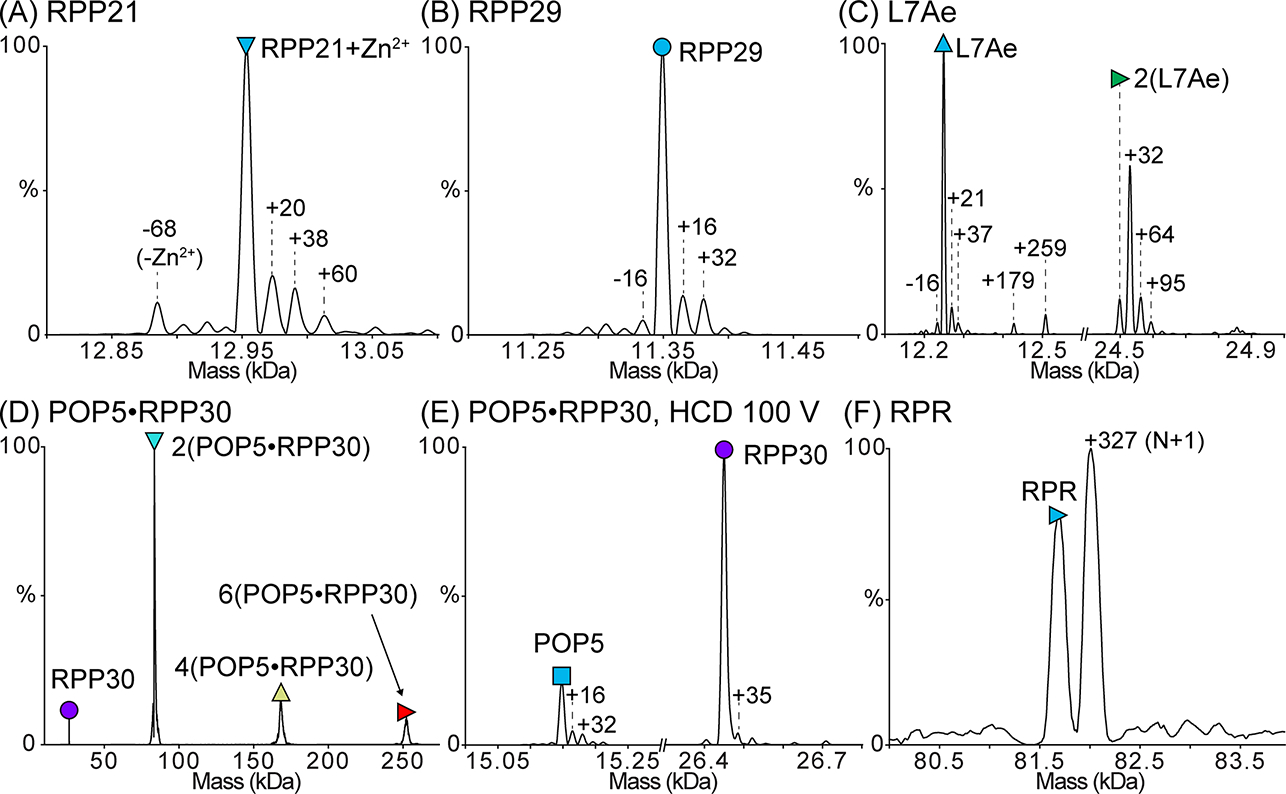
Proteins were dialyzed into 500 mM ammonium acetate prior to analysis. In each spectrum, the peak corresponding to the mass of the full-length subunit is indicated with a symbol. The expected and observed masses of each subunit are reported in Table 3. (A) Mass spectrum of RPP21 bound to Zn2+. Only a small population of RPP21 is Zn2+-free, indicating that Zn2+ remains tightly bound to RPP21 during all purification steps. (B) Mass spectrum of RPP29. (C) Mass spectrum of L7Ae. L7Ae readily forms a disulfide-mediated homodimer, which is lost upon treatment with DTT. The single-Cys-mediated dimer is also supported by the inability to dissociate it using HCD. (D) Mass spectrum of POP5·RPP30. This binary complex forms the expected heterotetramer [2(POP5·RPP30)] in addition to some higher-order oligomers. (E) Dissociation of the heterotetramer in (D) yields species with the expected masses for POP5 and RPP30. (F) Mass spectrum of RPR in 500 mM ammonium acetate and 0.4 mM Mg(OAc)2. Both the target RNA species (N) and a species extended by a single nucleotide (N+1), which is typical of T7 RNAP IVT, are observed. The mass spectrum confirms that the RPR is a mix of N and N+1 species with few contaminants (if any), suggesting that the doublet observed in a 2% (w/v) agarose gel likely results from alternative RPR conformations (see Note 13). In (A-E), mass differences (in Da) from the expected peak are indicated, and these differences may result from salt adducts or modifications, such as oxidation. Some of the adducted and/or modified peaks appear to have peak overlap as indicated by the peak asymmetry.
