Figure 5. Oligomerization of M. maripaludis POP5·RPP30 analyzed by SDS-PAGE and mass photometry.
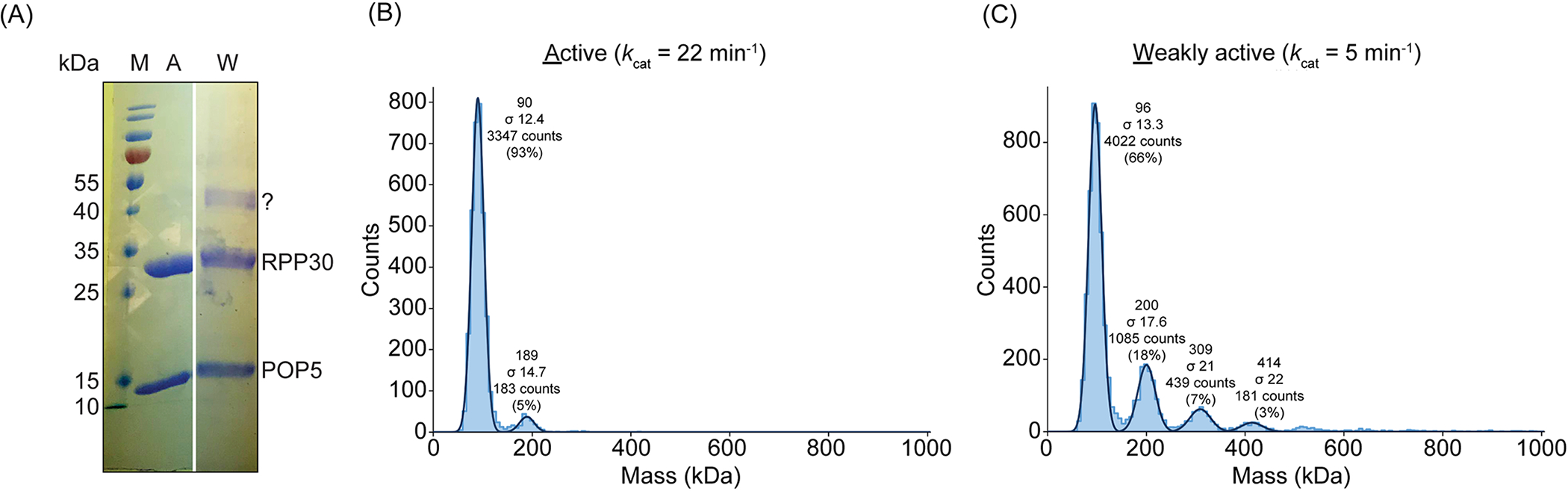
(A) SDS-PAGE (15% [w/v] polyacrylamide) analysis to compare highly active (H) and weakly active (W) preparations of POP5·RPP30. The gel image was spliced to position lanes of interest adjacent to each other. Although there is essentially no difference between the two preparations in the mobilities and relative intensities of the POP5 and RPP30 bands, the weakly active preparation has bands that migrate between the 40 and 55 kDa markers (M), likely corresponding to a heterodimer given the expected molecular weight of POP5·RPP30. The same two preparations of POP5·RPP30 (150 nM each) were diluted in 1X assay buffer (see 2.5) and analyzed by mass photometry (B and C). Histogram plotting and fitting was performed in DiscoverMP. The average mass of each population (in kDa), determined by the Gaussian fits, are indicated above each peak. For mass calculation, the following calibrants were used: lens epithelium-derived growth factor, LEDGF (60.4 kDa); bovine serum albumin, BSA (68.5 kDa); HIV integrase (80.4 kDa, monomer; 160.8 kDa, dimer); dihydroorotate dehydrogenase B (237 kDa); apoferritin (480 kDa); thyroglobulin (670 kDa). (B) Mass photometry analysis of a highly active preparation of POP5·RPP30. The major species (93%) corresponds to the POP5·RPP30 heterotetramer, which is the expected oligomeric state. A minor population of octamer (5%) is also observed. (C) Mass photometry analysis of a weakly active preparation of POP5·RPP30. While the major species is the heterotetramer (66%), the remaining species (29%) are octamer or larger.
